A DNA condensation code for linker histones
- PMID: 39116133
- PMCID: PMC11331069
- DOI: 10.1073/pnas.2409167121
A DNA condensation code for linker histones
Abstract
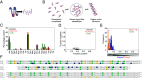
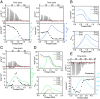
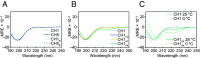
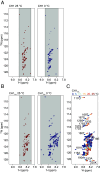
Linker histones play an essential role in chromatin packaging by facilitating compaction of the 11-nm fiber of nucleosomal "beads on a string." The result is a heterogeneous condensed state with local properties that range from dynamic, irregular, and liquid-like to stable and regular structures (the 30-nm fiber), which in turn impact chromatin-dependent activities at a fundamental level. The properties of the condensed state depend on the type of linker histone, particularly on the highly disordered C-terminal tail, which is the most variable region of the protein, both between species, and within the various subtypes and cell-type specific variants of a given organism. We have developed an in vitro model system comprising linker histone tail and linker DNA, which although very minimal, displays surprisingly complex behavior, and is sufficient to model the known states of linker histone-condensed chromatin: disordered "fuzzy" complexes ("open" chromatin), dense liquid-like assemblies (dynamic condensates), and higher-order structures (organized 30-nm fibers). A crucial advantage of such a simple model is that it allows the study of the various condensed states by NMR, circular dichroism, and scattering methods. Moreover, it allows capture of the thermodynamics underpinning the transitions between states through calorimetry. We have leveraged this to rationalize the distinct condensing properties of linker histone subtypes and variants across species that are encoded by the amino acid content of their C-terminal tails. Three properties emerge as key to defining the condensed state: charge density, lysine/arginine ratio, and proline-free regions, and we evaluate each separately using a strategic mutagenesis approach.
Keywords: chromatin; complex coacervation; intrinsically disordered protein; linker histone; phase separation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
-
- Gilbert N., Allan J., The many length scales of DNA packaging. Essays Biochem. 63, 13–16 (2019). - PubMed
-
- Kornberg R. D., Lorch Y., Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999). - PubMed
-
- Bednar J., Hamiche A., Dimitrov S., H1-nucleosome interactions and their functional implications. Biochim. Biophys. Acta 1859, 436–443 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

