A molecular pathway for cancer cachexia-induced muscle atrophy revealed at single-nucleus resolution
- PMID: 39116208
- PMCID: PMC11472345
- DOI: 10.1016/j.celrep.2024.114587
A molecular pathway for cancer cachexia-induced muscle atrophy revealed at single-nucleus resolution
Abstract
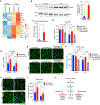
Cancer cachexia is a prevalent and often fatal wasting condition that cannot be fully reversed with nutritional interventions. Muscle atrophy is a central component of the syndrome, but the mechanisms whereby cancer leads to skeletal muscle atrophy are not well understood. We performed single-nucleus multi-omics on skeletal muscles from a mouse model of cancer cachexia and profiled the molecular changes in cachexic muscle. Our results revealed the activation of a denervation-dependent gene program that upregulates the transcription factor myogenin. Further studies showed that a myogenin-myostatin pathway promotes muscle atrophy in response to cancer cachexia. Short hairpin RNA inhibition of myogenin or inhibition of myostatin through overexpression of its endogenous inhibitor follistatin prevented cancer cachexia-induced muscle atrophy in mice. Our findings uncover a molecular basis of muscle atrophy associated with cancer cachexia and highlight potential therapeutic targets for this disorder.
Keywords: AAV; CP: Cancer; CP: Metabolism; atrophy; cachexia; denervation; myogenin; myostatin; single nucleus ATAC-seq; single nucleus RNA-seq; single nucleus multiome.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Evans WK, Makuch R, Clamon GH, Feld R, Weiner RS, Moran E, Blum R, Shepherd FA, Jeejeebhoy KN, and DeWys WD (1985). Limited impact of total parenteral nutrition on nutritional status during treatment for small cell lung cancer. Cancer Res. 45, 3347–3353. - PubMed
-
- Gannavarapu BS, Lau SKM, Carter K, Cannon NA, Gao A, Ahn C, Meyer JJ, Sher DJ, Jatoi A, Infante R, and Iyengar P (2018). Prevalence and Survival Impact of Pretreatment Cancer-Associated Weight Loss: A Tool for Guiding Early Palliative Care. J. Oncol. Pract. 14, e238–e250. 10.1200/JOP.2017.025221. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

