PTBP3 Mediates IL-18 Exon Skipping to Promote Immune Escape in Gallbladder Cancer
- PMID: 39116343
- PMCID: PMC11481411
- DOI: 10.1002/advs.202406633
PTBP3 Mediates IL-18 Exon Skipping to Promote Immune Escape in Gallbladder Cancer
Abstract
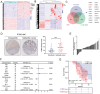
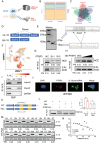
Gallbladder cancer (GBC) is the most common malignant tumor of the biliary system, with poor response to current treatments. Abnormal alternative splicing has been associated with the development of a variety of tumors. Combining the GEO database and GBC mRNA-seq analysis, it is found high expression of the splicing factor polypyrimidine region- binding protein 3 (PTBP3) in GBC. Multi-omics analysis revealed that PTBP3 promoted exon skipping of interleukin-18 (IL-18), resulting in the expression of ΔIL-18, an isoform specifically expressed in tumors. That ΔIL-18 promotes GBC immune escape by down-regulating FBXO38 transcription levels in CD8+T cells to reduce PD-1 ubiquitin-mediated degradation is revealed. Using a HuPBMC mouse model, the role of PTBP3 and ΔIL-18 in promoting GBC growth is confirmed, and showed that an antisense oligonucleotide that blocked ΔIL-18 production displayed anti-tumor activity. Furthermore, that the H3K36me3 promotes exon skipping of IL-18 by recruiting PTBP3 via MRG15 is demonstrated, thereby coupling the processes of IL-18 transcription and alternative splicing. Interestingly, it is also found that the H3K36 methyltransferase SETD2 binds to hnRNPL, thereby interfering with PTBP3 binding to IL-18 pre-mRNA. Overall, this study provides new insights into how aberrant alternative splicing mechanisms affect immune escape, and provides potential new perspectives for improving GBC immunotherapy.
Keywords: IL‐18; PTBP3; alternative splicing; gallbladder cancer; immune escape.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Yang Z.‐Y., Zhao C., Liu S.‐L., Pan L.‐J., Zhu Y.‐D., Zhao J.‐W., Wang H.‐K., Ye Y.‐Y., Qiang J., Shi L.‐Q., Mei J.‐W., Xie Y., Gong W., Shu Y.‐J., Dong P., Xiang S.‐S., Cancer Lett. 2024, 587, 216703. - PubMed
-
- Li Y., et al., Hepatology 2023, 78, 1352. - PubMed
-
- Zhao C., Zhang J., Yang Z.‐Y., Shi L.‐Q., Liu S.‐L., Pan L.‐J., Dong P., Zhang Y., Xiang S.‐S., Shu Y.‐J., Mei J.‐W., Phytomedicine 2023, 114, 154785. - PubMed
MeSH terms
Substances
Grants and funding
- 82172628/National Natural Science Foundation of China
- 82173081/National Natural Science Foundation of China
- 32201058/National Natural Science Foundation of China
- 82173048/National Natural Science Foundation of China
- 82373370/National Natural Science Foundation of China
- SACA-CY23C11/Shanghai Anticancer Association EYAS PROJECT
- 2022YQ002/Talent Program of Shanghai Health Care Commission
- 22Y11908000/Shanghai Scientific and Technological Innovation Plan
- 2022XD010/Shanghai Municipal Health Commission
- YG2024QNA19/Interdisciplinary Program of Shanghai Jiao Tong University
- YG2022ZD009/Interdisciplinary Program of Shanghai Jiao Tong University
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
