EGCG drives gut microbial remodeling-induced epithelial GPR43 activation to lessen Th1 polarization in colitis
- PMID: 39116526
- PMCID: PMC11363845
- DOI: 10.1016/j.redox.2024.103291
EGCG drives gut microbial remodeling-induced epithelial GPR43 activation to lessen Th1 polarization in colitis
Abstract
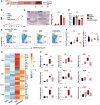
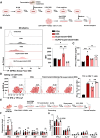
Modulation of immune microenvironment is critical for inflammatory bowel disease (IBD) intervention. Epigallocatechin gallate (EGCG), as a natural low toxicity product, has shown promise in treating IBD. However, whether and how EGCG regulates the intestinal microenvironment is not fully understood. Here we report that EGCG lessens colitis by orchestrating Th1 polarization and self-amplification in a novel manner that required multilevel-regulated intestinal microecosystem. Mechanistically, EGCG activates GPR43 on IEC to inhibit Th1 polarization dependently of short chain fatty acid (SCFA)-producing gut microbiota. Inhibition of GPR43 activity weakens the protective effects of EGCG on colitis development. Moreover, we confirm that fecal SCFAs and/or intestinal GPR43 are limited in patients with colitis and are correlated with Th1 cell number. Taken together, our study reveals an intestinal microenvironment-dependent immunoregulatory effects of EGCG in treating IBD and provides insight into mechanisms of EGCG-based novel immunotherapeutic strategies for IBD.
Keywords: EGCG; GPR43; Inflammatory bowel disease; Short chain fatty acids; Th1 cells.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Pan X., Zhu Q., Pan L.L., Sun J. Macrophage immunometabolism in inflammatory bowel diseases: from pathogenesis to therapy. Pharmacol. Therapeut. 2022;238:108176. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

