PanIN and CAF transitions in pancreatic carcinogenesis revealed with spatial data integration
- PMID: 39116880
- PMCID: PMC11409191
- DOI: 10.1016/j.cels.2024.07.001
PanIN and CAF transitions in pancreatic carcinogenesis revealed with spatial data integration
Abstract
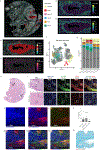
This study introduces a new imaging, spatial transcriptomics (ST), and single-cell RNA-sequencing integration pipeline to characterize neoplastic cell state transitions during tumorigenesis. We applied a semi-supervised analysis pipeline to examine premalignant pancreatic intraepithelial neoplasias (PanINs) that can develop into pancreatic ductal adenocarcinoma (PDAC). Their strict diagnosis on formalin-fixed and paraffin-embedded (FFPE) samples limited the single-cell characterization of human PanINs within their microenvironment. We leverage whole transcriptome FFPE ST to enable the study of a rare cohort of matched low-grade (LG) and high-grade (HG) PanIN lesions to track progression and map cellular phenotypes relative to single-cell PDAC datasets. We demonstrate that cancer-associated fibroblasts (CAFs), including antigen-presenting CAFs, are located close to PanINs. We further observed a transition from CAF-related inflammatory signaling to cellular proliferation during PanIN progression. We validate these findings with single-cell high-dimensional imaging proteomics and transcriptomics technologies. Altogether, our semi-supervised learning framework for spatial multi-omics has broad applicability across cancer types to decipher the spatiotemporal dynamics of carcinogenesis.
Keywords: Visium; Xenium; imaging mass cytometry; machine learning; multi-omics; pancreatic adenocarcinoma; pancreatic intraepithelial neoplasia; spatial transcriptomics; transfer learning.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.M.J. reports other support from Abmeta; personal fees from Genocea; personal fees from Achilles; personal fees from DragonFly; personal fees from Candel Therapeutics; other support from the Parker Institute; grants and other support from Lustgarten; personal fees from Carta; grants and other support from Genentech; grants and other support from AstraZeneca; personal fees from NextCure; and grants and other support from Break Through Cancer outside of the submitted work. E.J.F. is on the Scientific Advisory Board of Viosera Therapeutics/Resistance Bio and is a consultant to Mestag Therapeutics. W.J.H. reports patent royalties from Rodeo/Amgen and speaking/travel honoraria from Exelixis and Standard BioTools.
Figures





References
-
- Steele NG, Carpenter ES, Kemp SB, Sirihorachai VR, The S, Delrosario L, Lazarus J, Amir E-AD, Gunchick V, Espinoza C, et al. (2020). Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat. Cancer 1, 1097–1112. 10.1038/s43018-020-00121-4. - DOI - PMC - PubMed
-
- Lin W, Noel P, Borazanci EH, Lee J, Amini A, Han IW, Heo JS, Jameson GS, Fraser C, Steinbach M, et al. (2020). Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 12, 80. 10.1186/s13073-020-00776-9. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

