TorsinA is essential for neuronal nuclear pore complex localization and maturation
- PMID: 39117796
- PMCID: PMC11542706
- DOI: 10.1038/s41556-024-01480-1
TorsinA is essential for neuronal nuclear pore complex localization and maturation
Abstract
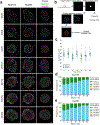
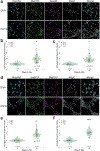
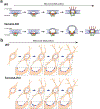
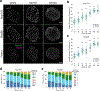
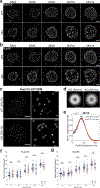
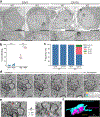
As lifelong interphase cells, neurons face an array of unique challenges. A key challenge is regulating nuclear pore complex (NPC) biogenesis and localization, the mechanisms of which are largely unknown. Here we identify neuronal maturation as a period of strongly upregulated NPC biogenesis. We demonstrate that the AAA+ protein torsinA, whose dysfunction causes the neurodevelopmental movement disorder DYT-TOR1A dystonia and co-ordinates NPC spatial organization without impacting total NPC density. We generated an endogenous Nup107-HaloTag mouse line to directly visualize NPC organization in developing neurons and find that torsinA is essential for proper NPC localization. In the absence of torsinA, the inner nuclear membrane buds excessively at sites of mislocalized nascent NPCs, and the formation of complete NPCs is delayed. Our work demonstrates that NPC spatial organization and number are independently determined and identifies NPC biogenesis as a process vulnerable to neurodevelopmental disease insults.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests:
None.
Figures


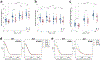






Update of
-
TorsinA is essential for the timing and localization of neuronal nuclear pore complex biogenesis.bioRxiv [Preprint]. 2023 Apr 27:2023.04.26.538491. doi: 10.1101/2023.04.26.538491. bioRxiv. 2023. Update in: Nat Cell Biol. 2024 Sep;26(9):1482-1495. doi: 10.1038/s41556-024-01480-1. PMID: 37162852 Free PMC article. Updated. Preprint.
References
Methods-only References
MeSH terms
Substances
Grants and funding
- R01 NS077730/NS/NINDS NIH HHS/United States
- 1S10OD021784/U.S. Department of Health & Human Services | NIH | NIH Office of the Director (OD)
- R01 NS110853/NS/NINDS NIH HHS/United States
- NS110853/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- S10 OD021784/OD/NIH HHS/United States
- R01 GM082949/GM/NIGMS NIH HHS/United States
- U24 NS120055/NS/NINDS NIH HHS/United States
- GM129347/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- GM007315/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 NS113943/NS/NINDS NIH HHS/United States
- R01 NS097542/NS/NINDS NIH HHS/United States
- MCB1552439/National Science Foundation (NSF)
- R56 NS128110/NS/NINDS NIH HHS/United States
- R01 GM129347/GM/NIGMS NIH HHS/United States
- NS128110/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- DK118480/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- NS113943/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 DK118480/DK/NIDDK NIH HHS/United States
- NSF2014862-UTA20-000890/National Science Foundation (NSF)
- NS077730/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- T32 GM007315/GM/NIGMS NIH HHS/United States
- NS097542/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

