Spermidine is essential for fasting-mediated autophagy and longevity
- PMID: 39117797
- PMCID: PMC11392816
- DOI: 10.1038/s41556-024-01468-x
Spermidine is essential for fasting-mediated autophagy and longevity
Abstract
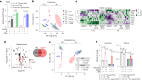
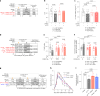
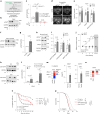
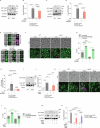
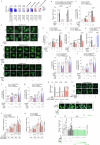
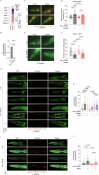
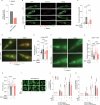
Caloric restriction and intermittent fasting prolong the lifespan and healthspan of model organisms and improve human health. The natural polyamine spermidine has been similarly linked to autophagy enhancement, geroprotection and reduced incidence of cardiovascular and neurodegenerative diseases across species borders. Here, we asked whether the cellular and physiological consequences of caloric restriction and fasting depend on polyamine metabolism. We report that spermidine levels increased upon distinct regimens of fasting or caloric restriction in yeast, flies, mice and human volunteers. Genetic or pharmacological blockade of endogenous spermidine synthesis reduced fasting-induced autophagy in yeast, nematodes and human cells. Furthermore, perturbing the polyamine pathway in vivo abrogated the lifespan- and healthspan-extending effects, as well as the cardioprotective and anti-arthritic consequences of fasting. Mechanistically, spermidine mediated these effects via autophagy induction and hypusination of the translation regulator eIF5A. In summary, the polyamine-hypusination axis emerges as a phylogenetically conserved metabolic control hub for fasting-mediated autophagy enhancement and longevity.
© 2024. The Author(s).
Conflict of interest statement
O.K., G.K. and F.M. are cofounders of Samsara Therapeutics, a company that develops novel pharmacological autophagy inducers. F.M. and T.E. have equity interests in and are advisors of The Longevity Labs. G.K. is a scientific co-founder of everImmune, Osasuna Therapeutics and Therafast Bio. G.K. has been holding research contracts with Daiichi Sankyo, Eleor, Kaleido, Lytix Pharma, PharmaMar, Osasuna Therapeutics, Samsara Therapeutics, Sanofi, Tollys and Vascage. G.K. is in the scientific advisory boards of Hevolution, Institut Servier and Rejuveron Life Sciences. F.G. and F.W.T. are employees of the Buchinger Wilhelmi Development und Holding, Überlingen. D.K., A.R.K. and A.M. are members of the steering committee of the Medical Association for Fasting and Nutrition (ÄGHE). D.K. and A.M. have also co-founded the Academy of Integrative Fasting, an institution for the qualification of medical staff in clinical fasting applications. D.K. serves as a consultant for a mobile application on intermittent fasting (FASTIC) as well as a company producing plant-based supplements (FENOU). A.M. is also a co-founder of the SALUFAST company and serves as a consultant for Lanserhof. All other authors declare no competing interests.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
- 310030_184781/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- Flagship project VASC-HEALTH/Medizinische Universität Graz (Medical University of Graz)
- TAI6021000/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- SI 849/14-1 (Project-ID 445178831)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- W1226/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- P27637-B28/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- I 3792/FWF_/Austrian Science Fund FWF/Austria
- P20 GM121176/GM/NIGMS NIH HHS/United States
- W 1226/FWF_/Austrian Science Fund FWF/Austria
- P 29203/FWF_/Austrian Science Fund FWF/Austria
- EXC2167-390884018/Deutsche Forschungsgemeinschaft (German Research Foundation)
- FOR2886/2-TP08/Deutsche Forschungsgemeinschaft (German Research Foundation)
- P33957/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- I3301-MINOTAUR/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- DOC-50/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- P29203/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- 310030_166474/184671/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- P 29262/FWF_/Austrian Science Fund FWF/Austria
- Field of Excellence BioHealth/Karl-Franzens-Universität Graz (University of Graz)
- P29262/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- P27893/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- F3012/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- P 27893/FWF_/Austrian Science Fund FWF/Austria
- 672032/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TAI 602/FWF_/Austrian Science Fund FWF/Austria
- P 33957/FWF_/Austrian Science Fund FWF/Austria
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

