Structural basis of RfaH-mediated transcription-translation coupling
- PMID: 39117885
- PMCID: PMC11927940
- DOI: 10.1038/s41594-024-01372-w
Structural basis of RfaH-mediated transcription-translation coupling
Abstract
The NusG paralog RfaH mediates bacterial transcription-translation coupling in genes that contain a DNA sequence element, termed an ops site, required for pausing RNA polymerase (RNAP) and for loading RfaH onto the paused RNAP. Here, we report cryo-electron microscopy structures of transcription-translation complexes (TTCs) containing Escherichia coli RfaH. The results show that RfaH bridges RNAP and the ribosome, with the RfaH N-terminal domain interacting with RNAP and the RfaH C-terminal domain interacting with the ribosome. The results show that the distribution of translational and orientational positions of RNAP relative to the ribosome in RfaH-coupled TTCs is more restricted than in NusG-coupled TTCs because of the more restricted flexibility of the RfaH interdomain linker. The results further suggest that the structural organization of RfaH-coupled TTCs in the 'loading state', in which RNAP and RfaH are located at the ops site during formation of the TTC, is the same as the structural organization of RfaH-coupled TTCs in the 'loaded state', in which RNAP and RfaH are located at positions downstream of the ops site during function of the TTC. The results define the structural organization of RfaH-containing TTCs and set the stage for analysis of functions of RfaH during translation initiation and transcription-translation coupling.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
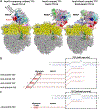
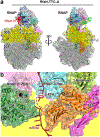
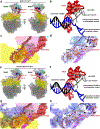

Update of
-
Structural basis of RfaH-mediated transcription-translation coupling.bioRxiv [Preprint]. 2023 Nov 6:2023.11.05.565726. doi: 10.1101/2023.11.05.565726. bioRxiv. 2023. Update in: Nat Struct Mol Biol. 2024 Dec;31(12):1932-1941. doi: 10.1038/s41594-024-01372-w. PMID: 37986937 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

