A defective splicing machinery promotes senescence through MDM4 alternative splicing
- PMID: 39118304
- PMCID: PMC11561654
- DOI: 10.1111/acel.14301
A defective splicing machinery promotes senescence through MDM4 alternative splicing
Abstract
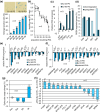
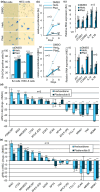
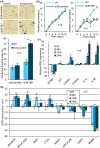
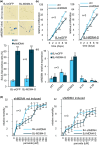
Defects in the splicing machinery are implicated in various diseases, including cancer. We observed a general reduction in the expression of spliceosome components and splicing regulators in human cell lines undergoing replicative, stress-induced, and telomere uncapping-induced senescence. Supporting the view that defective splicing contributes to senescence, splicing inhibitors herboxidiene, and pladienolide B induced senescence in normal and cancer cell lines. Furthermore, depleting individual spliceosome components also promoted senescence. All senescence types were associated with an alternative splicing transition from the MDM4-FL variant to MDM4-S. The MDM4 splicing shift was reproduced when splicing was inhibited, and spliceosome components were depleted. While decreasing the level of endogenous MDM4 promoted senescence and cell survival independently of the MDM4-S expression status, cell survival was also improved by increasing MDM4-S. Overall, our work establishes that splicing defects modulate the alternative splicing of MDM4 to promote senescence and cell survival.
Keywords: MDM4; RNA; alternative splicing; senescence; spliceosome; splicing factors.
© 2024 The Author(s). Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Acosta, J. C. , Banito, A. , Wuestefeld, T. , Georgilis, A. , Janich, P. , Morton, J. P. , Athineos, D. , Kang, T.‐W. , Lasitschka, F. , Andrulis, M. , Pascual, G. , Morris, K. J. , Khan, S. , Jin, H. , Dharmalingam, G. , Snijders, A. P. , Carroll, T. , Capper, D. , Pritchard, C. , … Gil, J. (2013). A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nature Cell Biology, 15(8), 978–990. 10.1038/ncb2784 - DOI - PMC - PubMed
-
- Allsopp, R. C. , Vaziri, H. , Patterson, C. , Goldstein, S. , Younglai, E. V. , Futcher, A. B. , Greider, C. W. , & Harley, C. B. (1992). Telomere length predicts replicative capacity of human fibroblasts. Proceedings of the National Academy of Sciences of the United States of America, 89(21), 10114–10118. - PMC - PubMed
-
- Alsafadi, S. , Houy, A. , Battistella, A. , Popova, T. , Wassef, M. , Henry, E. , Tirode, F. , Constantinou, A. , Piperno‐Neumann, S. , Roman‐Roman, S. , Dutertre, M. , & Stern, M.‐H. (2016). Cancer‐associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nature Communications, 7(1), 10615. 10.1038/ncomms10615 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

