Methuosis Inducer SGI-1027 Cooperates with Everolimus to Promote Apoptosis and Pyroptosis by Triggering Lysosomal Membrane Permeability in Renal Cancer
- PMID: 39119834
- PMCID: PMC11481186
- DOI: 10.1002/advs.202404693
Methuosis Inducer SGI-1027 Cooperates with Everolimus to Promote Apoptosis and Pyroptosis by Triggering Lysosomal Membrane Permeability in Renal Cancer
Abstract
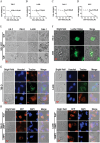
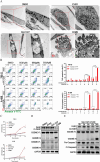
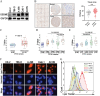
The mTOR inhibitor everolimus has been approved as a sequential or second-line therapy for renal cell carcinoma (RCC). However, the development of drug resistance limits its clinical applications. This study aims to address the challenge of everolimus resistance and provide new insights into the treatment of advanced RCC. Here, the cytotoxicity of the DNA methyltransferase 1 (DNMT1) inhibitor SGI-1027 in inducing cell vacuolation and methuosis is discovered and demonstrated for the first time. Additionally, SGI-1027 exerts synergistic effects with everolimus, as their combination suppresses the growth, migration, and invasion of renal cancer cells. Mechanistically, apoptosis and GSDME-dependent pyroptosis triggered by lysosomal membrane permeability (LMP) are observed. The upregulation of GSDME expression and increased lysosomal activity in renal cancer cells provide a therapeutic window for the combination of these two drugs to treat renal cancer. The combination treatment exhibits effective anti-tumor activity and is well tolerated in a subcutaneous tumor model. Overall, this study validates and reveals the specific cytotoxicity property of SGI-1027 and its potent synergistic effect with everolimus, offering new insights into advanced RCC therapy and everolimus-resistance overcoming.
Keywords: SGI‐1027; everolimus; lysosome; methuosis; pyroptosis; renal cell carcinoma.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Bukavina L., Bensalah K., Bray F., Carlo M., Challacombe B., Karam J. A., Kassouf W., Mitchell T., Montironi R., O'Brien T., Panebianco V., Scelo G., Shuch B., van Poppel H., Blosser C. D., Psutka S. P., Eur. Urol. 2022, 82, 529. - PubMed
-
- Klatte T., Rossi S. H., Stewart G. D., World J. Urol. 2018, 36, 1943. - PubMed
-
- Dudani S., de Velasco G., Wells J. C., Gan C. L., Donskov F., Porta C., Fraccon A., Pasini F., Lee J. L., Hansen A., Bjarnason G. A., Beuselinck B., Pal S. K., Yuasa T., Kroeger N., Kanesvaran R., Reaume M. N., Canil C., Choueiri T. K., Heng D. Y. C., JAMA Network Open 2021, 4, e2021869. - PMC - PubMed
-
- Lam J. S., Shvarts O., Leppert J. T., Pantuck A. J., Figlin R. A., Belldegrun A. S., J. Urol. 2005, 174, 466. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
