SEC16A Variants Predispose to Chronic Pancreatitis by Impairing ER-to-Golgi Transport and Inducing ER Stress
- PMID: 39119875
- PMCID: PMC11481239
- DOI: 10.1002/advs.202402550
SEC16A Variants Predispose to Chronic Pancreatitis by Impairing ER-to-Golgi Transport and Inducing ER Stress
Abstract
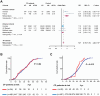
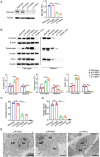
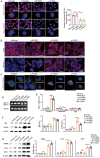
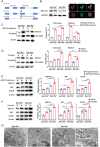
Chronic pancreatitis (CP) is a complex disease with genetic and environmental factors at play. Through trio exome sequencing, a de novo SEC16A frameshift variant in a Chinese teenage CP patient is identified. Subsequent targeted next-generation sequencing of the SEC16A gene in 1,061 Chinese CP patients and 1,196 controls reveals a higher allele frequency of rare nonsynonymous SEC16A variants in patients (4.90% vs 2.93%; odds ratio [OR], 1.71; 95% confidence interval [CI], 1.26-2.33). Similar enrichments are noted in a French cohort (OR, 2.74; 95% CI, 1.67-4.50) and in a biobank meta-analysis (OR, 1.16; 95% CI, 1.04-1.31). Notably, Chinese CP patients with SEC16A variants exhibit a median onset age 5 years earlier than those without (40.0 vs 45.0; p = 0.012). Functional studies using three CRISPR/Cas9-edited HEK293T cell lines show that loss-of-function SEC16A variants disrupt coat protein complex II (COPII) formation, impede secretory protein vesicles trafficking, and induce endoplasmic reticulum (ER) stress due to protein overload. Sec16a+/- mice, which demonstrate impaired zymogen secretion and exacerbated ER stress compared to Sec16a+/+, are further generated. In cerulein-stimulated pancreatitis models, Sec16a+/- mice display heightened pancreatic inflammation and fibrosis compared to wild-type mice. These findings implicate a novel pathogenic mechanism predisposing to CP.
Keywords: SEC16A; disease predisposition; endoplasmic reticulum stress; pancreatitis; protein vesicle trafficking.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Beyer G., Habtezion A., Werner J., Lerch M. M., Mayerle J., Lancet 2020, 396, 499. - PubMed
-
- Whitcomb D. C., Gorry M. C., Preston R. A., Furey W., Sossenheimer M. J., Ulrich C. D., Martin S. P., Gates L. K., Amann S. T., Toskes P. P., Liddle R., McGrath K., Uomo G., Post J. C., Ehrlich G. D., Nat. Genet. 1996, 14, 141. - PubMed
MeSH terms
Substances
Grants and funding
- 82222012/National Natural Science Foundation of China
- 81873588/National Natural Science Foundation of China
- 82120108006/National Natural Science Foundation of China
- 32070732/National Natural Science Foundation of China
- 201901070007E00052/Scientific Innovation Program of the Shanghai Municipal Education Committee
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
