PRMT6 Epigenetically Drives Metabolic Switch from Fatty Acid Oxidation toward Glycolysis and Promotes Osteoclast Differentiation During Osteoporosis
- PMID: 39120025
- PMCID: PMC11516099
- DOI: 10.1002/advs.202403177
PRMT6 Epigenetically Drives Metabolic Switch from Fatty Acid Oxidation toward Glycolysis and Promotes Osteoclast Differentiation During Osteoporosis
Abstract
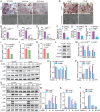
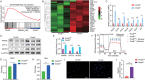
Epigenetic regulation of metabolism profoundly influences cell fate commitment. During osteoclast differentiation, the activation of RANK signaling is accompanied by metabolic reprogramming, but the epigenetic mechanisms by which RANK signaling induces this reprogramming remain elusive. By transcriptional sequence and ATAC analysis, this study identifies that activation of RANK signaling upregulates PRMT6 by epigenetic modification, triggering a metabolic switching from fatty acids oxidation toward glycolysis. Conversely, Prmt6 deficiency reverses this shift, markedly reducing HIF-1α-mediated glycolysis and enhancing fatty acid oxidation. Consequently, PRMT6 deficiency or inhibitor impedes osteoclast differentiation and alleviates bone loss in ovariectomized (OVX) mice. At the molecular level, Prmt6 deficiency reduces asymmetric dimethylation of H3R2 at the promoters of genes including Ppard, Acox3, and Cpt1a, enhancing genomic accessibility for fatty acid oxidation. PRMT6 thus emerges as a metabolic checkpoint, mediating metabolic switch from fatty acid oxidation to glycolysis, thereby supporting osteoclastogenesis. Unveiling PRMT6's critical role in epigenetically orchestrating metabolic shifts in osteoclastogenesis offers a promising target for anti-resorptive therapy.
Keywords: PRMT6; fatty acids oxidation; glycolysis; metabolic reprogramming; osteoclastogenesis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- McDonald M. M., Khoo W. H., Ng P. Y., Xiao Y., Zamerli J., Thatcher P., Kyaw W., Pathmanandavel K., Grootveld A. K., Moran I., Butt D., Nguyen A., Corr A., Warren S., Biro M., Butterfield N. C., Guilfoyle S. E., Komla‐Ebri D., Dack M. R. G., Dewhurst H. F., Logan J. G., Li Y., Mohanty S. T., Byrne N., Terry R. L., Simic M. K., Chai R., Quinn J. M. W., Youlten S. E., Pettitt J. A., et al., Cell 2021, 184, 1940. - PMC - PubMed
-
- Novack D. V., Teitelbaum S. L., Annu. Rev. Pathol. 2008, 3, 457. - PubMed
-
- Park S., Heo J. S., Mizuno S., Kim M., An H., Hong E., Kang M. G., Kim J., Yun R., Park H., Noh E. H., Lee M. J., Yoon K., Kim P., Son M., Pang K., Lee J., Park J., Ooshima A., Kim T. J., Park J. Y., Yang K. M., Myung S. J., Bae H., Lee K. M., Letterio J., Park S. H., Takahashi S., Kim S. J., Metabolism 2023, 151, 155746. - PubMed
-
- Eastell R., O'Neill T. W., Hofbauer L. C., Langdahl B., Reid I. R., Gold D. T., Cummings S. R., Nat. Rev. Dis. Primers 2016, 2, 16069. - PubMed
-
- Boyle W. J., Simonet W. S., Lacey D. L., Nature 2003, 423, 337. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
