Two cyanobacterial species exhibit stress responses when grown together in visible light or far-red light
- PMID: 39120135
- PMCID: PMC11423583
- DOI: 10.1128/msphere.00251-24
Two cyanobacterial species exhibit stress responses when grown together in visible light or far-red light
Abstract
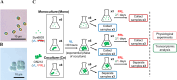
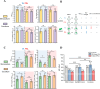
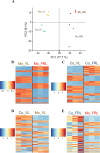
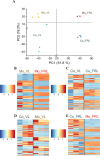
Although most cyanobacteria grow in visible light (VL; λ = 400-700 nm), some cyanobacteria can also use far-red light (FRL; λ = 700-800 nm) for oxygenic photosynthesis by performing far-red light photoacclimation. These two types of cyanobacteria can be found in the same environment. However, how they respond to each other remains unknown. Here, we reveal that coculture stresses FRL-using Chlorogloeopsis fritschii PCC 9212 and VL-using Synechocystis sp. PCC 6803. No significant growth difference was found in Synechocystis sp. PCC 6803 between the coculture and the monoculture. Conversely, the growth of Chlorogloeopsis fritschii PCC 9212 was suppressed in VL under coculture. According to transcriptomic analysis, Chlorogloeopsis fritschii PCC 9212 in coculture shows low transcript levels of metabolic activities and high transcript levels of ion transporters, with the differences being more noticeable in VL than in FRL. The transcript levels of stress responses in coculture were likewise higher than in monoculture in Synechocystis sp. PCC 6803 under FRL. The low transcript level of metabolic activities in coculture or the inhibition of cyanobacterial growth indicates a possible negative interaction between these two cyanobacterial strains.IMPORTANCEThe interaction between two cyanobacterial species is the primary focus of this study. One species harvests visible light, while the other can harvest far-red and visible light. Prior research on cyanobacteria interaction concentrated on its interactions with algal, coral, and fungal species. Interactions between cyanobacterial species were, nevertheless, rarely discussed. Thus, we characterized the interaction between two cyanobacterial species, one capable of photosynthesis using far-red light and the other not. Through experimental and bioinformatic approaches, we demonstrate that when one cyanobacterium thrives under optimal light conditions, it stresses the remaining cyanobacterial species. We contribute to an ecological understanding of these two kinds of cyanobacteria distribution patterns. Cyanobacteria that utilize far-red light probably disperse in environments with limited visible light to avoid competition with other cyanobacteria. From a biotechnological standpoint, this study suggests that the simultaneous cultivation of two cyanobacterial species in large-scale cultivation facilities may reduce the overall biomass yield.
Keywords: coculture; cyanobacteria; far-red light photoacclimation (FaRLiP); negative interaction; transcriptomic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sinha RP, Häder D-P. 2008. UV-protectants in cyanobacteria. Plant Sci 174:278–289. doi:10.1016/j.plantsci.2007.12.004 - DOI
-
- Ho M-Y, Bryant DA. 2021. Photosynthesis | long wavelength pigments in photosynthesis, p 245–255. In Jez J (ed), Encyclopedia of biological chemistry III, 3rd ed. Elsevier, Oxford.
-
- McCree KJ. 1973. The measurement of photosynthetically active radiation. Solar Energy 15:83–87. doi:10.1016/0038-092X(73)90010-8 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
