Synthetic mRNAs Containing Minimalistic Untranslated Regions Are Highly Functional In Vitro and In Vivo
- PMID: 39120273
- PMCID: PMC11311775
- DOI: 10.3390/cells13151242
Synthetic mRNAs Containing Minimalistic Untranslated Regions Are Highly Functional In Vitro and In Vivo
Abstract
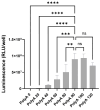
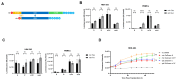
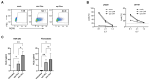
Synthetic mRNA produced by in vitro transcription (ivt mRNA) is the active pharmaceutical ingredient of approved anti-COVID-19 vaccines and of many drugs under development. Such synthetic mRNA typically contains several hundred bases of non-coding "untranslated" regions (UTRs) that are involved in the stabilization and translation of the mRNA. However, UTRs are often complex structures, which may complicate the entire production process. To eliminate this obstacle, we managed to reduce the total amount of nucleotides in the UTRs to only four bases. In this way, we generate minimal ivt mRNA ("minRNA"), which is less complex than the usual optimized ivt mRNAs that are contained, for example, in approved vaccines. We have compared the efficacy of minRNA to common augmented mRNAs (with UTRs of globin genes or those included in licensed vaccines) in vivo and in vitro and could demonstrate equivalent functionalities. Our minimal mRNA design will facilitate the further development and implementation of ivt mRNA-based vaccines and therapies.
Keywords: UTR; in vitro transcription; ivt mRNA; mRNA; untranslated regions.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

