Investigating Splice Defects in USH2A Using Targeted Long-Read Sequencing
- PMID: 39120292
- PMCID: PMC11311777
- DOI: 10.3390/cells13151261
Investigating Splice Defects in USH2A Using Targeted Long-Read Sequencing
Abstract
Biallelic variants in USH2A are associated with retinitis pigmentosa (RP) and Type 2 Usher Syndrome (USH2), leading to impaired vision and, additionally, hearing loss in the latter. Although the introduction of next-generation sequencing into clinical diagnostics has led to a significant uplift in molecular diagnostic rates, many patients remain molecularly unsolved. It is thought that non-coding variants or variants of uncertain significance contribute significantly to this diagnostic gap. This study aims to demonstrate the clinical utility of the reverse transcription-polymerase chain reaction (RT-PCR)-Oxford Nanopore Technology (ONT) sequencing of USH2A mRNA transcripts from nasal epithelial cells to determine the splice-altering effect of candidate variants. Five affected individuals with USH2 or non-syndromic RP who had undergone whole genome sequencing were recruited for further investigation. All individuals had uncertain genotypes in USH2A, including deep intronic rare variants, c.8682-654C>G, c.9055+389G>A, and c.9959-2971C>T; a synonymous variant of uncertain significance, c.2139C>T; p.(Gly713=); and a predicted loss of function duplication spanning an intron/exon boundary, c.3812-3_3837dup p.(Met1280Ter). In silico assessment using SpliceAI provided splice-altering predictions for all candidate variants which were investigated using ONT sequencing. All predictions were found to be accurate; however, in the case of c.3812-3_3837dup, the outcome was a complex cryptic splicing pattern with predominant in-frame exon 18 skipping and a low level of exon 18 inclusion leading to the predicted stop gain. This study detected and functionally characterised simple and complex mis-splicing patterns in USH2A arising from previously unknown deep intronic variants and previously reported variants of uncertain significance, confirming the pathogenicity of the variants.
Keywords: Oxford nanopore sequencing; USH2A; cryptic splice-altering variant; deep intronic variant; inherited retinal disease; long-read sequencing; nasal epithelial cells.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
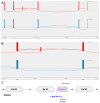
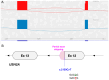


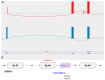
References
-
- Von Graefe A. Vereizelte Beobachtungen und Bemerkungen. Exceptionnelles verhalter des Gesichts feldes bei Pigmentenarter der Netzhalt. Arch. Klin. Ophtalmol. 1858;4:250–253.
-
- Hufnagel R.B., Liang W., Duncan J.L., Brewer C.C., Audo I., Ayala A.R., Branham K., Cheetham J.K., Daiger S.P., Durham T.A., et al. Tissue-specific genotype-phenotype correlations among USH2A-related disorders in the RUSH2A study. Hum. Mutat. 2022;43:613–624. doi: 10.1002/humu.24365. - DOI - PMC - PubMed
-
- Lenassi E., Vincent A., Li Z., Saihan Z., Coffey A.J., Steele-Stallard H.B., Moore A.T., Steel K.P., Luxon L.M., Héon E., et al. A detailed clinical and molecular survey of subjects with nonsyndromic USH2A retinopathy reveals an allelic hierarchy of disease-causing variants. Eur. J. Hum. Genet. 2015;23:1318–1327. doi: 10.1038/ejhg.2014.283. - DOI - PMC - PubMed
-
- Lin S., Vermeirsch S., Pontikos N., Martin-Gutierrez M.P., Daich Varela M., Malka S., Schiff E., Knight H., Wright G., Jurkute N., et al. Spectrum of Genetic Variants in the Most Common Genes Causing Inherited Retinal Disease in a Large Molecularly Characterized United Kingdom Cohort. Ophthalmol. Retina. 2024;12:699–709. doi: 10.1016/j.oret.2024.01.012. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 206619/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- 5045/46/Fight for Sight UK
- Stephen and Elizabeth Archer in memory of Marion Woods/Moorfields Eye Charity
- N/A/National Institute of Health Research Biomedical Research Centre (NIHR-BRC) at Moorfields Eye Hospital and UCL Institute of Ophthalmology
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

