MAPK Signaling-Mediated RFNG Phosphorylation and Nuclear Translocation Restrain Oxaliplatin-Induced Apoptosis and Ferroptosis
- PMID: 39120977
- PMCID: PMC11481204
- DOI: 10.1002/advs.202402795
MAPK Signaling-Mediated RFNG Phosphorylation and Nuclear Translocation Restrain Oxaliplatin-Induced Apoptosis and Ferroptosis
Abstract
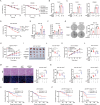
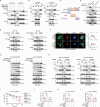
Chemotherapy resistance remains a major challenge in the treatment of colorectal cancer (CRC). Therefore, it is crucial to develop novel strategies to sensitize cancer cells to chemotherapy. Here, the fringe family is screened to determine their contribution to chemotherapy resistance in CRC. It is found that RFNG depletion significantly sensitizes cancer cells to oxaliplatin treatment. Mechanistically, chemotherapy-activated MAPK signaling induces ERK to phosphorylate RFNG Ser255 residue. Phosphorylated RFNG S255 (pS255) interacts with the nuclear importin proteins KPNA1/importin-α1 and KPNB1/importin-β1, leading to its translocation into the nucleus where it targets p53 and inhibits its phosphorylation by competitively inhibiting the binding of CHK2 to p53. Consequently, the expression of CDKN1A is decreased and that of SLC7A11 is increased, leading to the inhibition of apoptosis and ferroptosis. In contrast, phosphor-deficient RFNG S225A mutant showed increased apoptosis and ferroptosis, and exhibited a notable response to oxaliplatin chemotherapy both in vitro and in vivo. It is further revealed that patients with low RFNG pS255 exhibited significant sensitivity to oxaliplatin in a patient-derived xenograft (PDX) model. These findings highlight the crosstalk between the MAPK and p53 signaling pathways through RFNG, which mediates oxaliplatin resistance in CRC. Additionally, this study provides guidance for oxaliplatin treatment of CRC patients.
Keywords: MAPK signaling; apoptosis and ferroptosis; nuclear RFNG; oxaliplatin chemosensitivity; p53 activity.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Woloshin S., Kramer B. S., N. Engl. J. Med. 2022, 387, 91. - PubMed
-
- Petrelli F., Labianca R., Zaniboni A., Lonardi S., Galli F., Rulli E., Rosati G., Corallo S., Ronzoni M., Cardellino G. G., Mattioli R., Mambrini A., Ciuffreda L., Banzi M., Pusceddu V., Maiello E., Zampino M., Zagonel V., Marchetti P., Corsi D., Rimassa L., Cinieri S., Sobrero A., JAMA Oncol. 2020, 6, 547. - PMC - PubMed
-
- de Gramont A., Figer A., Seymour M., Homerin M., Hmissi A., Cassidy J., Boni C., Cortes‐Funes H., Cervantes A., Freyer G., Papamichael D., Bail N. L., Louvet C., Hendler D., de Braud F., Wilson C., Morvan F., Bonetti A., J. Clin. Oncol. 2000, 18, 2938. - PubMed
-
- Crea F., Nobili S., Paolicchi E., Perrone G., Napoli C., Landini I., Danesi R., Mini E., Drug Resist. Updat. 2011, 14, 280. - PubMed
-
- Levine A. J., Nat. Rev. Cancer 2020, 20, 471. - PubMed
MeSH terms
Substances
Grants and funding
- 82203152/National Natural Science Foundation of China
- 82303614/National Natural Science Foundation of China
- 92359302/National Natural Science Foundation of China
- 82303923/National Natural Science Foundation of China
- 2022A1515111079/Guangdong Basic and Applied Basic Research Foundation
- 2021B1515230009/Guangdong Basic and Applied Basic Research Foundation
- 2022YFC3401000/National Key Research And Development Plan
- 2023B1111020005/Guangdong Provincial Key Areas R&D Programs of "Precision medicine and stem cells"
- 2022T150760/China Postdoctoral Science Foundation
- 2023T160750/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
