Effects of storage conditions on the microbiota of fecal samples collected from dairy cattle
- PMID: 39121104
- PMCID: PMC11315314
- DOI: 10.1371/journal.pone.0308571
Effects of storage conditions on the microbiota of fecal samples collected from dairy cattle
Abstract
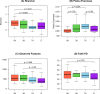
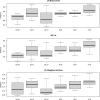
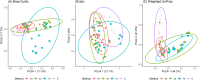
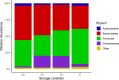
Microbiota analyses are key to understanding the bacterial communities within dairy cattle, but the impact of different storage conditions on these analyses remains unclear. This study sought to examine the effects of freezing at -80°C immediately after collection, refrigeration at 4°C for three days and seven days and absolute ethanol preservation on the microbiota diversity of pooled fecal samples from dairy cattle. Examining 16S rRNA gene sequences, alpha (Shannon, Pielou evenness, observed features and Faith PD indices) and beta (Bray-Curtis, βw and Weighted UniFrac) diversity were assessed. The effects of storage conditions on these metrics were evaluated using linear mixed models and PERMANOVA, incorporating the farm as a random effect. Our findings reveal that 7d and E significantly altered the Shannon index, suggesting a change in community composition. Changes in Pielou evenness for 3d and 7d storage when compared to 0d were found, indicating a shift in species evenness. Ethanol preservation impacted both observed features and Faith PD indices. Storage conditions significantly influenced Bray-Curtis, βw, and Weighted UniFrac metrics, indicating changes in community structure. PERMANOVA analysis showed that these storage conditions significantly contributed to microbiota differences compared to immediate freezing. In conclusion, our results demonstrate that while refrigeration for three days had minimal impact, seven days of refrigeration and ethanol preservation significantly altered microbiota analyses. These findings highlight the importance of sample storage considerations in microbiota research.
Copyright: © 2024 Jaramillo-Jaramillo et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Assessment of the impact of different fecal storage protocols on the microbiota diversity and composition: a pilot study.BMC Microbiol. 2019 Jun 28;19(1):145. doi: 10.1186/s12866-019-1519-2. BMC Microbiol. 2019. PMID: 31253096 Free PMC article.
-
Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles.BMC Microbiol. 2017 Mar 31;17(1):78. doi: 10.1186/s12866-017-0983-9. BMC Microbiol. 2017. PMID: 28359329 Free PMC article.
-
Comparison of fecal and oral collection methods for studies of the human microbiota in two Iranian cohorts.BMC Microbiol. 2021 Nov 22;21(1):324. doi: 10.1186/s12866-021-02387-9. BMC Microbiol. 2021. PMID: 34809575 Free PMC article.
-
Collection media and delayed freezing effects on microbial composition of human stool.Microbiome. 2015 Aug 12;3:33. doi: 10.1186/s40168-015-0092-7. eCollection 2015. Microbiome. 2015. PMID: 26269741 Free PMC article.
-
Effects of Stool Sample Preservation Methods on Gut Microbiota Biodiversity: New Original Data and Systematic Review with Meta-Analysis.Microbiol Spectr. 2023 Jun 15;11(3):e0429722. doi: 10.1128/spectrum.04297-22. Epub 2023 Apr 24. Microbiol Spectr. 2023. PMID: 37093040 Free PMC article.
References
-
- Nazir A. Review on Metagenomics and its Applications. Imp J Interdiscip Res. 2016. Jan 1;2. ISSN: 2454-1362.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

