IL-1 receptor antagonism reveals a yin-yang relationship between NFκB and interferon signaling in chronic lymphocytic leukemia
- PMID: 39121163
- PMCID: PMC11331101
- DOI: 10.1073/pnas.2405644121
IL-1 receptor antagonism reveals a yin-yang relationship between NFκB and interferon signaling in chronic lymphocytic leukemia
Abstract
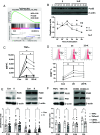
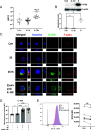
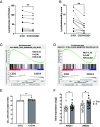
Nuclear factor kappa B (NFκB) is a pathogenic factor in chronic lymphocytic leukemia (CLL) that is not addressed specifically by current therapies. NFκB is activated by inflammatory factors that stimulate toll-like receptors (TLRs) and receptors for interleukin-1 (IL-1) family members. IL-1 is considered a master regulator of inflammation, and IL-1 receptor signaling is inhibited by the IL-1 receptor antagonist anakinra. These considerations suggested that anakinra might have a role in the treatment of CLL. Consistent with this idea, anakinra inhibited spontaneous and TLR7-mediated activation of the canonical NFκB pathway in CLL cells in vitro. However, CLL cells exhibited only weak signaling responses to IL-1 itself, and anakinra was found to inhibit NFκB along with oxidative stress in an IL-1 receptor-independent manner. Anakinra was then administered with minimal toxicity to 11 previously untreated CLL patients in a phase I dose-escalation trial (NCT04691765). A stereotyped clinical response was observed in all patients. Anakinra lowered blood lymphocytes and lymph node sizes within the first month that were associated with downregulation of NFκB and oxidative stress in the leukemia cells. However, inhibition of NFκB was accompanied by upregulation of type 1 interferon (IFN) signaling, c-MYC-regulated genes and proteins, and loss of the initial clinical response. Anakinra increased IFN signaling and survival of CLL cells in vitro that were, respectively, phenocopied by mitochondrial antioxidants and reversed by IFN receptor blocking antibodies. These observations suggest that anakinra has activity in CLL and may be a useful adjunct for conventional therapies as long as compensatory IFN signaling is blocked at the same time.
Keywords: IL1 receptor antagonist; chronic lymphocytic leukemia; interferon; signal transduction.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Fabbri G., Dalla-Favera R., The molecular pathogenesis of chronic lymphocytic leukaemia. Nat. Rev. Cancer 16, 145–162 (2016). - PubMed
-
- Martines C., et al. , Macrophage- and BCR-derived but not TLR-derived signals support the growth of CLL and Richter syndrome murine models in vivo. Blood 140, 2335–2347 (2022). - PubMed
-
- Mansouri L., Papakonstantinou N., Ntoufa S., Stamatopoulos K., Rosenquist R., NF-κB activation in chronic lymphocytic leukemia: A point of convergence of external triggers and intrinsic lesions. Semin. Cancer Biol. 39, 40–48 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

