Altered enhancer-promoter interaction leads to MNX1 expression in pediatric acute myeloid leukemia with t(7;12)(q36;p13)
- PMID: 39121370
- PMCID: PMC11460460
- DOI: 10.1182/bloodadvances.2023012161
Altered enhancer-promoter interaction leads to MNX1 expression in pediatric acute myeloid leukemia with t(7;12)(q36;p13)
Abstract
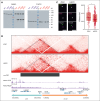
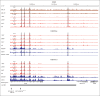
Acute myeloid leukemia (AML) with the t(7;12)(q36;p13) translocation occurs only in very young children and has a poor clinical outcome. The expected oncofusion between break point partners (motor neuron and pancreas homeobox 1 [MNX1] and ETS variant transcription factor 6 [ETV6]) has only been reported in a subset of cases. However, a universal feature is the strong transcript and protein expression of MNX1, a homeobox transcription factor that is normally not expressed in hematopoietic cells. Here, we map the translocation break points on chromosomes 7 and 12 in affected patients to a region proximal to MNX1 and either introns 1 or 2 of ETV6. The frequency of MNX1 overexpression in pediatric AML is 2.4% and occurs predominantly in t(7;12)(q36;p13) AML. Chromatin interaction assays in a t(7;12)(q36;p13) induced pluripotent stem cell line model unravel an enhancer-hijacking event that explains MNX1 overexpression in hematopoietic cells. Our data suggest that enhancer hijacking may be a more widespread consequence of translocations in which no oncofusion product was identified, including t(1;3) or t(4;12) AML.
© 2024 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: L.B. has received honoraria from AbbVie, Amgen, Astellas, Bristol Myers Squibb, Celgene, Daiichi Sankyo, Gilead, Hexal, Janssen, Jazz Pharmaceuticals, Menarini, Novartis, Pfizer, Roche, and Sanofi; and research support from Bayer and Jazz Pharmaceuticals. D.B.L. receives honoraria from Infectopharm GmbH. The remaining authors declare no competing financial interests.
Figures




References
-
- de Thé H, Pandolfi PP, Chen Z. Acute promyelocytic leukemia: a paradigm for oncoprotein-targeted cure. Cancer Cell. 2017;32(5):552–560. - PubMed
-
- Tosi S, Giudici G, Mosna G, et al. Identification of new partner chromosomes involved in fusions with the ETV6 (TEL) gene in hematologic malignancies. Genes Chromosomes Cancer. 1998;21(3):223–229. - PubMed
-
- Wlodarska I, La Starza R, Baens M, et al. Fluorescence in situ hybridization characterization of new translocations involving TEL (ETV6) in a wide spectrum of hematologic malignancies. Blood. 1998;91(4):1399–1406. - PubMed
-
- Espersen ADL, Noren-Nyström U, Abrahamsson J, et al. Acute myeloid leukemia (AML) with t(7;12)(q36;p13) is associated with infancy and trisomy 19: data from Nordic Society for Pediatric Hematology and Oncology (NOPHO-AML) and review of the literature. Genes Chromosomes Cancer. 2018;57(7):359–365. - PubMed
-
- von Bergh ARM, van Drunen E, van Wering ER, et al. High incidence of t(7;12)(q36;p13) in infant AML but not in infant ALL, with a dismal outcome and ectopic expression of HLXB9. Genes Chromosomes Cancer. 2006;45(8):731–739. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

