Epigenetic and Oncogenic Inhibitors Cooperatively Drive Differentiation and Kill KRAS-Mutant Colorectal Cancers
- PMID: 39121480
- PMCID: PMC11609823
- DOI: 10.1158/2159-8290.CD-23-0866
Epigenetic and Oncogenic Inhibitors Cooperatively Drive Differentiation and Kill KRAS-Mutant Colorectal Cancers
Abstract
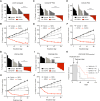
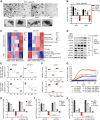
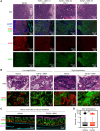
Combined EZH2 and RAS pathway inhibitors kill KRAS-mutant colorectal cancer cells and promote durable tumor regression in vivo. These agents function by cooperatively suppressing the WNT pathway, driving differentiation, and epigenetically reprogramming cells to permit the induction of apoptotic signals, which then kill these more differentiated tumor cells.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
P. Loi reports grants from the NIH/NCI during the conduct of the study. A.E. Schade reports grants from the American Cancer Society during the conduct of the study. M. Watanabe reports grants from Brigham and Women’s Hospital during the conduct of the study. O. Popow reports grants from Cancer Research UK and the Mark Foundation for Cancer Research during the conduct of the study. N. Gunduz reports grants from Cancer Grand Challenges during the conduct of the study. M. Giannakis reports grants from Janssen and personal fees from Nerviano Medical Sciences outside the submitted work. K. Ng reports nonfinancial support from Pharmavite, grants from Janssen, personal fees from Bayer, GlaxoSmithKline, Pfizer, CytomX, Jazz Pharmaceuticals, Revolution Medicines, AbbVie, Etiome, Seagen, CRICO, and JAMA outside the submitted work. S. Santagata reports being speaker honoraria for Novartis and Roche. K. Helin reports grants from the Cancer Research UK Grand Challenge and the Mark Foundation for Cancer Research to the SPECIFICANCER team during the conduct of the study. O.J. Sansom reports grants from AstraZeneca, Novartis, Boehringer Ingelheim, and Cancer Research Horizons outside the submitted work. K. Cichowski reports grants from Cancer Research UK and the Mark Foundation for Cancer Research during the conduct of the study, as well as other support from Erasca outside the submitted work. No disclosures were reported by the other authors.
Figures




References
-
- O’Connell JB, Maggard MB, Ko CY. Colon cancer survival rates with the new American joint committee on cancer sixth edition staging. J Natl Cancer Inst 2004;96:1420–5. - PubMed
-
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383:1490–502. - PubMed
-
- Morel D, Jeffery D, Aspeslagh S, Almouzni G, Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours—past lessons and future promise. Nat Rev Clin Oncol 2020;17:91–107. - PubMed
-
- Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol 2018;15:709–20. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

