Cdc14 phosphatases use an intramolecular pseudosubstrate motif to stimulate and regulate catalysis
- PMID: 39122012
- PMCID: PMC11407943
- DOI: 10.1016/j.jbc.2024.107644
Cdc14 phosphatases use an intramolecular pseudosubstrate motif to stimulate and regulate catalysis
Abstract
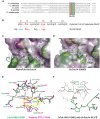
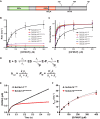
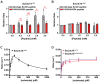
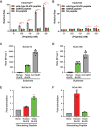
Cdc14 phosphatases are related structurally and mechanistically to protein tyrosine phosphatases (PTPs) but evolved a unique specificity for phosphoSer-Pro-X-Lys/Arg sites primarily deposited by cyclin-dependent kinases. This specialization is widely conserved in eukaryotes. The evolutionary reconfiguration of the Cdc14 active site to selectively accommodate phosphoSer-Pro likely required modification to the canonical PTP catalytic cycle. While studying Saccharomyces cerevisiae Cdc14, we discovered a short sequence in the disordered C terminus, distal to the catalytic domain, which mimics an optimal substrate. Kinetic analyses demonstrated this pseudosubstrate binds the active site and strongly stimulates rate-limiting phosphoenzyme hydrolysis, and we named it "substrate-like catalytic enhancer" (SLiCE). The SLiCE motif is found in all Dikarya fungal Cdc14 orthologs and contains an invariant glutamine, which we propose is positioned via substrate-like contacts to assist orientation of the hydrolytic water, similar to a conserved active site glutamine in other PTPs that Cdc14 lacks. AlphaFold2 predictions revealed vertebrate Cdc14 orthologs contain a conserved C-terminal alpha helix bound to the active site. Although apparently unrelated to the fungal sequence, this motif also makes substrate-like contacts and has an invariant glutamine in the catalytic pocket. Altering these residues in human Cdc14A and Cdc14B demonstrated that it functions by the same mechanism as the fungal motif. However, the fungal and vertebrate SLiCE motifs were not functionally interchangeable, illuminating potential active site differences during catalysis. Finally, we show that the fungal SLiCE motif is a target for phosphoregulation of Cdc14 activity. Our study uncovered evolution of an unusual stimulatory pseudosubstrate motif in Cdc14 phosphatases.
Keywords: Cdc14; enzyme catalysis; enzyme kinetics; enzyme mechanism; enzyme regulation; phosphatase; phosphorylation; pseudosubstrate; structural model.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
Kinetic and mechanistic studies of a cell cycle protein phosphatase Cdc14.J Biol Chem. 2004 Jul 16;279(29):30459-68. doi: 10.1074/jbc.M402217200. Epub 2004 May 5. J Biol Chem. 2004. PMID: 15128740
-
The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase.EMBO J. 2003 Jul 15;22(14):3524-35. doi: 10.1093/emboj/cdg348. EMBO J. 2003. PMID: 12853468 Free PMC article.
-
A Substrate Trapping Method for Identification of Direct Cdc14 Phosphatase Targets.Methods Mol Biol. 2017;1505:119-132. doi: 10.1007/978-1-4939-6502-1_10. Methods Mol Biol. 2017. PMID: 27826861
-
The diverging role of CDC14B: from mitotic exit in yeast to cell fate control in humans.EMBO J. 2023 Aug 15;42(16):e114364. doi: 10.15252/embj.2023114364. Epub 2023 Jul 26. EMBO J. 2023. PMID: 37493185 Free PMC article. Review.
-
Closing mitosis: the functions of the Cdc14 phosphatase and its regulation.Annu Rev Genet. 2004;38:203-32. doi: 10.1146/annurev.genet.38.072902.093051. Annu Rev Genet. 2004. PMID: 15568976 Review.
References
-
- Culotti J., Hartwell L.H. Genetic control of the cell division cycle in yeast. Exp. Cell Res. 1971;67:389–401. - PubMed
-
- Taylor G.S., Liu Y., Baskerville C., Charbonneau H. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J. Biol. Chem. 1997;272:24054–24063. - PubMed
-
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. - PubMed
-
- Mocciaro A., Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J. Cell Sci. 2010;123:2867–2876. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

