Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility
- PMID: 39122675
- PMCID: PMC11316121
- DOI: 10.1038/s41467-024-50930-9
Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility
Abstract
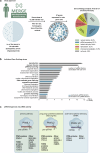
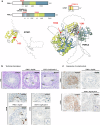
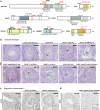
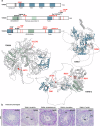
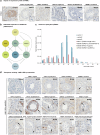
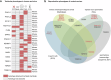
piRNAs are crucial for transposon silencing, germ cell maturation, and fertility in male mice. Here, we report on the genetic landscape of piRNA dysfunction in humans and present 39 infertile men carrying biallelic variants in 14 different piRNA pathway genes, including PIWIL1, GTSF1, GPAT2, MAEL, TDRD1, and DDX4. In some affected men, the testicular phenotypes differ from those of the respective knockout mice and range from complete germ cell loss to the production of a few morphologically abnormal sperm. A reduced number of pachytene piRNAs was detected in the testicular tissue of variant carriers, demonstrating impaired piRNA biogenesis. Furthermore, LINE1 expression in spermatogonia links impaired piRNA biogenesis to transposon de-silencing and serves to classify variants as functionally relevant. These results establish the disrupted piRNA pathway as a major cause of human spermatogenic failure and provide insights into transposon silencing in human male germ cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Comment in
-
piRNA pathway disruption in human infertility.Nat Rev Urol. 2024 Oct;21(10):577. doi: 10.1038/s41585-024-00946-z. Nat Rev Urol. 2024. PMID: 39271802 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

