Super enhancer acquisition drives expression of oncogenic PPP1R15B that regulates protein homeostasis in multiple myeloma
- PMID: 39122682
- PMCID: PMC11316114
- DOI: 10.1038/s41467-024-50910-z
Super enhancer acquisition drives expression of oncogenic PPP1R15B that regulates protein homeostasis in multiple myeloma
Abstract
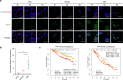
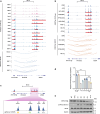
Multiple myeloma is a hematological malignancy arising from immunoglobulin-secreting plasma cells. It remains poorly understood how chromatin rewiring of regulatory elements contributes to tumorigenesis and therapy resistance in myeloma. Here we generate a high-resolution contact map of myeloma-associated super-enhancers by integrating H3K27ac ChIP-seq and HiChIP from myeloma cell lines, patient-derived myeloma cells and normal plasma cells. Our comprehensive transcriptomic and phenomic analyses prioritize candidate genes with biological and clinical implications in myeloma. We show that myeloma cells frequently acquire SE that transcriptionally activate an oncogene PPP1R15B, which encodes a regulatory subunit of the holophosphatase complex that dephosphorylates translation initiation factor eIF2α. Epigenetic silencing or knockdown of PPP1R15B activates pro-apoptotic eIF2α-ATF4-CHOP pathway, while inhibiting protein synthesis and immunoglobulin production. Pharmacological inhibition of PPP1R15B using Raphin1 potentiates the anti-myeloma effect of bortezomib. Our study reveals that myeloma cells are vulnerable to perturbation of PPP1R15B-dependent protein homeostasis, highlighting a promising therapeutic strategy.
© 2024. The Author(s).
Conflict of interest statement
M.J.F. declares two patents on methodologies related to chromatin interaction analysis by paired-end tag sequencing (ChIA-PET). The remaining authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

