Hydrogel crosslinking modulates macrophages, fibroblasts, and their communication, during wound healing
- PMID: 39122702
- PMCID: PMC11315930
- DOI: 10.1038/s41467-024-50072-y
Hydrogel crosslinking modulates macrophages, fibroblasts, and their communication, during wound healing
Abstract
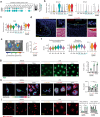
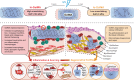
Biomaterial wound dressings, such as hydrogels, interact with host cells to regulate tissue repair. This study investigates how crosslinking of gelatin-based hydrogels influences immune and stromal cell behavior and wound healing in female mice. We observe that softer, lightly crosslinked hydrogels promote greater cellular infiltration and result in smaller scars compared to stiffer, heavily crosslinked hydrogels. Using single-cell RNA sequencing, we further show that heavily crosslinked hydrogels increase inflammation and lead to the formation of a distinct macrophage subpopulation exhibiting signs of oxidative activity and cell fusion. Conversely, lightly crosslinked hydrogels are more readily taken up by macrophages and integrated within the tissue. The physical properties differentially affect macrophage and fibroblast interactions, with heavily crosslinked hydrogels promoting pro-fibrotic fibroblast activity that drives macrophage fusion through RANKL signaling. These findings suggest that tuning the physical properties of hydrogels can guide cellular responses and improve healing, offering insights for designing better biomaterials for wound treatment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- P30AR075047/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- S10 OD025064/OD/NIH HHS/United States
- R01AR079150/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- 594598/Simons Foundation
- R01AR079470/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- R01 AI151301/AI/NIAID NIH HHS/United States
- S10OD025064/U.S. Department of Health & Human Services | NIH | NIH Office of the Director (OD)
- U01AR073159/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- DMS1763272/NSF | Directorate for Mathematical & Physical Sciences | Division of Mathematical Sciences (DMS)
- R01 AR079470/AR/NIAMS NIH HHS/United States
- 1R01AI151301/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- 1R21AR077288/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- R21 AR077288/AR/NIAMS NIH HHS/United States
- R01 AR079150/AR/NIAMS NIH HHS/United States
- U01 AR073159/AR/NIAMS NIH HHS/United States
- P30 AR075047/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

