The Defective in Autoregulation (DAR) gene of Medicago truncatula encodes a protein involved in regulating nodulation and arbuscular mycorrhiza
- PMID: 39123119
- PMCID: PMC11316349
- DOI: 10.1186/s12870-024-05479-6
The Defective in Autoregulation (DAR) gene of Medicago truncatula encodes a protein involved in regulating nodulation and arbuscular mycorrhiza
Erratum in
-
Correction: The Defective in Autoregulation (DAR) gene of Medicago truncatula encodes a protein involved in regulating nodulation and arbuscular mycorrhiza.BMC Plant Biol. 2025 Feb 14;25(1):193. doi: 10.1186/s12870-025-06215-4. BMC Plant Biol. 2025. PMID: 39953401 Free PMC article. No abstract available.
Abstract
Background: Legumes utilize a long-distance signaling feedback pathway, termed Autoregulation of Nodulation (AON), to regulate the establishment and maintenance of their symbiosis with rhizobia. Several proteins key to this pathway have been discovered, but the AON pathway is not completely understood.
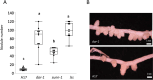
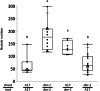
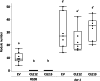
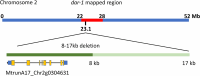
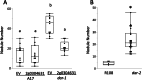
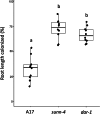
Results: We report a new hypernodulating mutant, defective in autoregulation, with disruption of a gene, DAR (Medtr2g450550/MtrunA17_Chr2g0304631), previously unknown to play a role in AON. The dar-1 mutant produces ten-fold more nodules than wild type, similar to AON mutants with disrupted SUNN gene function. As in sunn mutants, suppression of nodulation by CLE peptides MtCLE12 and MtCLE13 is abolished in dar. Furthermore, dar-1 also shows increased root length colonization by an arbuscular mycorrhizal fungus, suggesting a role for DAR in autoregulation of mycorrhizal symbiosis (AOM). However, unlike SUNN which functions in the shoot to control nodulation, DAR functions in the root.
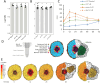
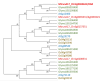
Conclusions: DAR encodes a membrane protein that is a member of a small protein family in M. truncatula. Our results suggest that DAR could be involved in the subcellular transport of signals involved in symbiosis regulation, but it is not upregulated during symbiosis. DAR gene family members are also present in Arabidopsis, lycophytes, mosses, and microalgae, suggesting the AON and AOM may use pathway components common to other plants, even those that do not undergo either symbiosis.
Keywords: Medicago truncatula; AOM; AON; Autoregulation of mycorrhizal symbiosis; Autoregulation of nodulation; DAR; Hypernodulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
CLE peptide tri-arabinosylation and peptide domain sequence composition are essential for SUNN-dependent autoregulation of nodulation in Medicago truncatula.New Phytol. 2018 Apr;218(1):73-80. doi: 10.1111/nph.15019. Epub 2018 Feb 2. New Phytol. 2018. PMID: 29393515
-
Unraveling new molecular players involved in the autoregulation of nodulation in Medicago truncatula.J Exp Bot. 2019 Feb 20;70(4):1407-1417. doi: 10.1093/jxb/ery465. J Exp Bot. 2019. PMID: 30753553 Free PMC article.
-
Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula.New Phytol. 2011 Jun;190(4):865-874. doi: 10.1111/j.1469-8137.2011.03738.x. Epub 2011 Apr 20. New Phytol. 2011. PMID: 21507004
-
Common and divergent roles of plant hormones in nodulation and arbuscular mycorrhizal symbioses.Plant Signal Behav. 2014;9(9):e29593. doi: 10.4161/psb.29593. Plant Signal Behav. 2014. PMID: 25763697 Free PMC article. Review.
-
Functional genomics dissection of the nodulation autoregulation pathway (AON) in soybean (Glycine max).J Integr Plant Biol. 2025 Mar;67(3):762-772. doi: 10.1111/jipb.13898. Epub 2025 Mar 24. J Integr Plant Biol. 2025. PMID: 40125797 Review.
Cited by
-
Functional Genomics of Legumes in Bulgaria-Advances and Future Perspectives.Genes (Basel). 2025 Feb 28;16(3):296. doi: 10.3390/genes16030296. Genes (Basel). 2025. PMID: 40149448 Free PMC article. Review.
-
Correction: The Defective in Autoregulation (DAR) gene of Medicago truncatula encodes a protein involved in regulating nodulation and arbuscular mycorrhiza.BMC Plant Biol. 2025 Feb 14;25(1):193. doi: 10.1186/s12870-025-06215-4. BMC Plant Biol. 2025. PMID: 39953401 Free PMC article. No abstract available.
References
-
- Oldroyd GE, Leyser O. A plant’s diet, surviving in a variable nutrient environment. Science. 2020;368(6486):eaba0196. - PubMed
-
- Ferguson BJ, Mens C, Hastwell AH, Zhang M, Su H, Jones CH, et al. Legume nodulation: the host controls the party. Plant, Cell Environ. 2019;42(1):41–51. - PubMed
-
- Pervent M, Lambert I, Tauzin M, Karouani A, Nigg M, Jardinaud M-F, et al. Systemic control of nodule formation by plant nitrogen demand requires autoregulation-dependent and independent mechanisms. J Exp Bot. 2021;72(22):7942–56. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

