Genomic landscape of hepatocellular carcinoma in Egyptian patients by whole exome sequencing
- PMID: 39123171
- PMCID: PMC11311965
- DOI: 10.1186/s12920-024-01965-w
Genomic landscape of hepatocellular carcinoma in Egyptian patients by whole exome sequencing
Erratum in
-
Correction: Genomic landscape of hepatocellular carcinoma in Egyptian patients by whole exome sequencing.BMC Med Genomics. 2024 Nov 12;17(1):268. doi: 10.1186/s12920-024-02045-9. BMC Med Genomics. 2024. PMID: 39533256 Free PMC article. No abstract available.
Abstract
Background: Hepatocellular carcinoma (HCC) is the most common primary liver cancer. Chronic hepatitis and liver cirrhosis lead to accumulation of genetic alterations driving HCC pathogenesis. This study is designed to explore genomic landscape of HCC in Egyptian patients by whole exome sequencing.
Methods: Whole exome sequencing using Ion Torrent was done on 13 HCC patients, who underwent surgical intervention (7 patients underwent living donor liver transplantation (LDLT) and 6 patients had surgical resection}.
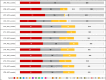
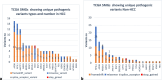
Results: Mutational signature was mostly S1, S5, S6, and S12 in HCC. Analysis of highly mutated genes in both HCC and Non-HCC revealed the presence of highly mutated genes in HCC (AHNAK2, MUC6, MUC16, TTN, ZNF17, FLG, MUC12, OBSCN, PDE4DIP, MUC5b, and HYDIN). Among the 26 significantly mutated HCC genes-identified across 10 genome sequencing studies-in addition to TCGA, APOB and RP1L1 showed the highest number of mutations in both HCC and Non-HCC tissues. Tier 1, Tier 2 variants in TCGA SMGs in HCC and Non-HCC (TP53, PIK3CA, CDKN2A, and BAP1). Cancer Genome Landscape analysis revealed Tier 1 and Tier 2 variants in HCC (MSH2) and in Non-HCC (KMT2D and ATM). For KEGG analysis, the significantly annotated clusters in HCC were Notch signaling, Wnt signaling, PI3K-AKT pathway, Hippo signaling, Apelin signaling, Hedgehog (Hh) signaling, and MAPK signaling, in addition to ECM-receptor interaction, focal adhesion, and calcium signaling. Tier 1 and Tier 2 variants KIT, KMT2D, NOTCH1, KMT2C, PIK3CA, KIT, SMARCA4, ATM, PTEN, MSH2, and PTCH1 were low frequency variants in both HCC and Non-HCC.
Conclusion: Our results are in accordance with previous studies in HCC regarding highly mutated genes, TCGA and specifically enriched pathways in HCC. Analysis for clinical interpretation of variants revealed the presence of Tier 1 and Tier 2 variants that represent potential clinically actionable targets. The use of sequencing techniques to detect structural variants and novel techniques as single cell sequencing together with multiomics transcriptomics, metagenomics will integrate the molecular pathogenesis of HCC in Egyptian patients.
Keywords: Hepatocelleular carcinoma (HCC); Living Donor Liver Transplantation (LDLT); Mutational signature; Whole exome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics: GLOBOCAN estimates of incidence andmortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Elghazaly H, GabAllah A, Eldin NB. P-019 Clinic-pathological pattern of hepatocellular carcinoma (HCC) in Egypt. Ann Oncol. 2018;29. 10.1093/annonc/mdy151.018.
-
- Wang H, Zhou H, Li X, Wang P, Liu G, Liu W, et al. Detection of tumor- related biomarkers in hepatocellular carcinoma patients by sequencing circulating cell-free DNA. Clin Oncol. 2019;4:1645.
-
- Maloberti T, De Leo A, Sanza V, Gruppioni E, Altimari A, Riefolo M, Visani M, Malvi D, D’Errico A, Tallini G, Vasuri F, de Biase D. Correlation of molecular alterations with pathological features in hepatocellular carcinoma: literature review and experience of an Italian center. World J Gastroenterol. 2022;28(25):2854–66. - PMC - PubMed
-
- European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C. J Hepatol. 2018;69(2):461–511. - PubMed
MeSH terms
Grants and funding
- Basic and Applied Research Grant Call 7 (BARG Call 7, Project ID:38229)./Science and Technology Development Fund
- Basic and Applied Research Grant Call 7 (BARG Call 7, Project ID:38229)./Science and Technology Development Fund
- Basic and Applied Research Grant Call 7 (BARG Call 7, Project ID:38229)./Science and Technology Development Fund
- Basic and Applied Research Grant Call 7 (BARG Call 7, Project ID:38229)./Science and Technology Development Fund
- Basic and Applied Research Grant Call 7 (BARG Call 7, Project ID:38229)./Science and Technology Development Fund
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

