Autophagy-mediated ID1 turnover dictates chemo-resistant fate in ovarian cancer stem cells
- PMID: 39123206
- PMCID: PMC11316295
- DOI: 10.1186/s13046-024-03147-z
Autophagy-mediated ID1 turnover dictates chemo-resistant fate in ovarian cancer stem cells
Abstract
Background: The mechanisms enabling dynamic shifts between drug-resistant and drug-sensitive states in cancer cells are still underexplored. This study investigated the role of targeted autophagic protein degradation in regulating ovarian cancer stem cell (CSC) fate decisions and chemo-resistance.
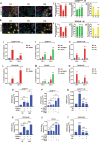
Methods: Autophagy levels were compared between CSC-enriched side population (SP) and non-SP cells (NSP) in multiple ovarian cancer cell lines using immunoblotting, immunofluorescence, and transmission electron microscopy. The impact of autophagy modulation on CSC markers and differentiation was assessed by flow cytometry, immunoblotting and qRT-PCR. In silico modeling and co-immunoprecipitation identified ID1 interacting proteins. Pharmacological and genetic approaches along with Annexin-PI assay, ChIP assay, western blotting, qRT-PCR and ICP-MS were used to evaluate effects on cisplatin sensitivity, apoptosis, SLC31A1 expression, promoter binding, and intracellular platinum accumulation in ID1 depleted backdrop. Patient-derived tumor spheroids were analyzed for autophagy and SLC31A1 levels.
Results: Ovarian CSCs exhibited increased basal autophagy compared to non-CSCs. Further autophagy stimulation by serum-starvation and chemical modes triggered proteolysis of the stemness regulator ID1, driving the differentiation of chemo-resistant CSCs into chemo-sensitive non-CSCs. In silico modeling predicted TCF12 as a potent ID1 interactor, which was validated by co-immunoprecipitation. ID1 depletion freed TCF12 to transactivate the cisplatin influx transporter SLC31A1, increasing intracellular cisplatin levels and cytotoxicity. Patient-derived tumor spheroids exhibited a functional association between autophagy, ID1, SLC31A1, and platinum sensitivity.
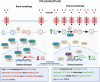
Conclusions: This study reveals a novel autophagy-ID1-TCF12-SLC31A1 axis where targeted autophagic degradation of ID1 enables rapid remodeling of CSCs to reverse chemo-resistance. Modulating this pathway could counter drug resistance in ovarian cancer.
Keywords: Autophagy; Cancer stem cells; Chemo-resistance; Differentiation; Epithelial Ovarian Cancer; ID1; SLC31A1; TCF12.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

