Phylogenetic and Genetic Variation Analysis of Porcine Epidemic Diarrhea Virus in East Central China during 2020-2023
- PMID: 39123710
- PMCID: PMC11311003
- DOI: 10.3390/ani14152185
Phylogenetic and Genetic Variation Analysis of Porcine Epidemic Diarrhea Virus in East Central China during 2020-2023
Abstract
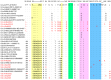
Porcine epidemic diarrhea virus (PEDV) is a major causative pathogen of a highly contagious, acute enteric viral disease. This study evaluated the emergence of nine variants in Jiangsu and Anhui provinces of China from 2020 to 2023. S gene-based phylogenetic analysis indicated that three variants belong to the G1c subgroup, while the other six strains are clustered within the G2c subgroup. Recombination analyses supported that three variants of the G1c subgroup were likely derived from recombination of parental variants FR0012014 and a donor variant AJ1102. In addition, there are novel mutations on amino acid 141-148 and these likely resulted in changes in antigenicity in the three variants. These results illustrated that the study provides novel insights into the epidemiology, evolution, and transmission of PEDV in China.
Keywords: PEDV; Spike gene; antigenic analysis; phylogenetic analysis; recombination analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Molecular characterization of porcine epidemic diarrhea virus associated with outbreaks in southwest China during 2014-2018.Transbound Emerg Dis. 2021 Nov;68(6):3482-3497. doi: 10.1111/tbed.13953. Epub 2021 Feb 7. Transbound Emerg Dis. 2021. PMID: 33306274
-
Isolation and Characterization of Porcine Epidemic Diarrhea Virus G2c Strains Circulating in China from 2021 to 2024.Vet Sci. 2025 May 6;12(5):444. doi: 10.3390/vetsci12050444. Vet Sci. 2025. PMID: 40431537 Free PMC article.
-
Detection and phylogenetic analysis of porcine epidemic diarrhea virus in central China based on the ORF3 gene and the S1 gene.Virol J. 2016 Nov 25;13(1):192. doi: 10.1186/s12985-016-0646-8. Virol J. 2016. PMID: 27887624 Free PMC article.
-
Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains.Virus Res. 2016 Dec 2;226:20-39. doi: 10.1016/j.virusres.2016.05.023. Epub 2016 Jun 8. Virus Res. 2016. PMID: 27288724 Free PMC article. Review.
-
Epidemiology of porcine epidemic diarrhea virus among Chinese pig populations: A meta-analysis.Microb Pathog. 2019 Apr;129:43-49. doi: 10.1016/j.micpath.2019.01.017. Epub 2019 Jan 23. Microb Pathog. 2019. PMID: 30682525
Cited by
-
Engineering a recombination-resistant live attenuated vaccine candidate with suppressed interferon antagonists for PEDV.J Virol. 2025 Jul 22;99(7):e0045125. doi: 10.1128/jvi.00451-25. Epub 2025 Jun 12. J Virol. 2025. PMID: 40503881 Free PMC article.
-
Antiviral Activity of 1-Deoxynojirimycin Extracts of Mulberry Leaves Against Porcine Epidemic Diarrhea Virus.Animals (Basel). 2025 Apr 23;15(9):1207. doi: 10.3390/ani15091207. Animals (Basel). 2025. PMID: 40362022 Free PMC article.
-
Genetic characterization and pathogenicity analysis of three porcine epidemic diarrhea virus strains isolated from North China.Vet Res. 2025 Jun 14;56(1):118. doi: 10.1186/s13567-025-01554-4. Vet Res. 2025. PMID: 40517255 Free PMC article.
References
-
- He W.T., Bollen N., Xu Y., Zhao J., Dellicour S., Yan Z., Gong W., Zhang C., Zhang L., Lu M., et al. Phylogeography Reveals Association between Swine Trade and the Spread of Porcine Epidemic Diarrhea Virus in China and across the World. Mol. Biol. Evol. 2022;39:msab364. doi: 10.1093/molbev/msab364. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

