Structural Unfolding of G-Quadruplexes: From Small Molecules to Antisense Strategies
- PMID: 39124893
- PMCID: PMC11314335
- DOI: 10.3390/molecules29153488
Structural Unfolding of G-Quadruplexes: From Small Molecules to Antisense Strategies
Abstract
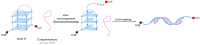
G-quadruplexes (G4s) are non-canonical nucleic acid secondary structures that have gathered significant interest in medicinal chemistry over the past two decades due to their unique structural features and potential roles in a variety of biological processes and disorders. Traditionally, research efforts have focused on stabilizing G4s, while in recent years, the attention has progressively shifted to G4 destabilization, unveiling new therapeutic perspectives. This review provides an in-depth overview of recent advances in the development of small molecules, starting with the controversial role of TMPyP4. Moreover, we described effective metal complexes in addition to G4-disrupting small molecules as well as good G4 stabilizing ligands that can destabilize G4s in response to external stimuli. Finally, we presented antisense strategies as a promising approach for destabilizing G4s, with a particular focus on 2'-OMe antisense oligonucleotide, peptide nucleic acid, and locked nucleic acid. Overall, this review emphasizes the importance of understanding G4 dynamics as well as ongoing efforts to develop selective G4-unfolding strategies that can modulate their biological function and therapeutic potential.
Keywords: G-clamp; G-quadruplex; antisense strategy; disrupting small molecules; locked nucleic acid; new therapeutic strategies.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures








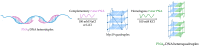



Similar articles
-
Structurally diverse G-quadruplexes as the noncanonical nucleic acid drug target for live cell imaging and antibacterial study.Chem Commun (Camb). 2023 Feb 2;59(11):1415-1433. doi: 10.1039/d2cc05945b. Chem Commun (Camb). 2023. PMID: 36636928 Review.
-
Identifying G-Quadruplex-DNA-Disrupting Small Molecules.J Am Chem Soc. 2021 Aug 18;143(32):12567-12577. doi: 10.1021/jacs.1c04426. Epub 2021 Aug 4. J Am Chem Soc. 2021. PMID: 34346684
-
G-Quadruplexes in the Viral Genome: Unlocking Targets for Therapeutic Interventions and Antiviral Strategies.Viruses. 2023 Nov 5;15(11):2216. doi: 10.3390/v15112216. Viruses. 2023. PMID: 38005893 Free PMC article. Review.
-
Stability of Human Telomeric G-Quadruplexes Complexed with Photosensitive Ligands and Irradiated with Visible Light.Int J Mol Sci. 2023 May 22;24(10):9090. doi: 10.3390/ijms24109090. Int J Mol Sci. 2023. PMID: 37240437 Free PMC article.
-
Topologies of G-quadruplex: Biological functions and regulation by ligands.Biochem Biophys Res Commun. 2020 Oct 8;531(1):3-17. doi: 10.1016/j.bbrc.2019.12.103. Epub 2020 Jan 14. Biochem Biophys Res Commun. 2020. PMID: 31948752 Review.
Cited by
-
RNA G-quadruplex structure targeting and imaging: recent advances and future directions.RNA. 2025 Jul 16;31(8):1053-1080. doi: 10.1261/rna.080587.125. RNA. 2025. PMID: 40473419 Free PMC article. Review.
-
RNA G-quadruplexes: emerging regulators of gene expression and therapeutic targets.Funct Integr Genomics. 2025 Jul 3;25(1):143. doi: 10.1007/s10142-025-01656-4. Funct Integr Genomics. 2025. PMID: 40608121 Review.
-
Myriad factors and pathways influencing tumor radiotherapy resistance.Open Life Sci. 2024 Nov 26;19(1):20220992. doi: 10.1515/biol-2022-0992. eCollection 2024. Open Life Sci. 2024. PMID: 39655194 Free PMC article. Review.
-
Fluorescence Detection of DNA/RNA G-Quadruplexes (G4s) by Twice-as-Smart Ligands.ChemMedChem. 2025 Apr 1;20(7):e202400829. doi: 10.1002/cmdc.202400829. Epub 2025 Jan 15. ChemMedChem. 2025. PMID: 39714851 Free PMC article.
-
Untargeted CUT&Tag reads are enriched at accessible chromatin and restrict identification of potential G4-forming sequences in G4-targeted CUT&Tag experiments.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf678. doi: 10.1093/nar/gkaf678. Nucleic Acids Res. 2025. PMID: 40682824 Free PMC article.
References
-
- Zuffo M., Pirota V., Doria F. Photochemistry: Volume 46. Volume 46. The Royal Society of Chemistry; London, UK: 2019. Photoresponsive molecular devices targeting nucleic acid secondary structures; pp. 281–318. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

