An Optimized Peptide Antagonist of CXCR4 Limits Survival of BCR-ABL1-Transformed Cells in Philadelphia-Chromosome-Positive B-Cell Acute Lymphoblastic Leukemia
- PMID: 39125877
- PMCID: PMC11312813
- DOI: 10.3390/ijms25158306
An Optimized Peptide Antagonist of CXCR4 Limits Survival of BCR-ABL1-Transformed Cells in Philadelphia-Chromosome-Positive B-Cell Acute Lymphoblastic Leukemia
Abstract
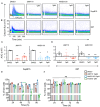
Philadelphia-chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) is characterized by reciprocal chromosomal translocation between chromosome 9 and 22, leading to the expression of constitutively active oncogenic BCR-ABL1 fusion protein. CXC chemokine receptor 4 (CXCR4) is essential for the survival of BCR-ABL1-transformed mouse pre-B cells, as the deletion of CXCR4 induces death in these cells. To investigate whether CXCR4 inhibition also effectively blocks BCR-ABL1-transformed cell growth in vitro, in this study, we explored an array of peptide-based inhibitors of CXCR4. The inhibitors were optimized derivatives of EPI-X4, an endogenous peptide antagonist of CXCR4. We observed that among all the candidates, EPI-X4 JM#170 (referred to as JM#170) effectively induced cell death in BCR-ABL1-transformed mouse B cells but had little effect on untransformed wild-type B cells. Importantly, AMD3100, a small molecule inhibitor of CXCR4, did not show this effect. Treatment with JM#170 induced transient JNK phosphorylation in BCR-ABL1-transformed cells, which in turn activated the intrinsic apoptotic pathway by inducing cJun, Bim, and Bax gene expressions. Combinatorial treatment of JM#170 with ABL1 kinase inhibitor Imatinib exerted a stronger killing effect on BCR-ABL1-transformed cells even at a lower dose of Imatinib. Surprisingly, JM#170 actively killed Sup-B15 cells, a BCR-ABL1+ human ALL cell line, but had no effect on the BCR-ABL1- 697 cell line. This suggests that the inhibitory effect of JM#170 is specific for BCR-ABL1+ ALL. Taken together, JM#170 emerges as a potent novel drug against Ph+ ALL.
Keywords: AMD3100; BCR–ABL1; CXCR4; EPI-X4 derivatives; Imatinib; cell survival.
Conflict of interest statement
M.H. and J.M. are co-inventors of pending and issued patents that claim to use EPI-X4 (ALB408-423) and its derivatives for the therapy of CXCR4-associated diseases.
Figures
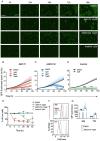

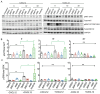
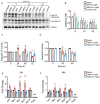
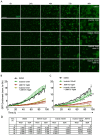
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

