Neutrophil-like Monocytes Increase in Patients with Colon Cancer and Induce Dysfunctional TIGIT+ NK Cells
- PMID: 39126041
- PMCID: PMC11313383
- DOI: 10.3390/ijms25158470
Neutrophil-like Monocytes Increase in Patients with Colon Cancer and Induce Dysfunctional TIGIT+ NK Cells
Abstract
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous family of immune cells including granulocytic (CD14neg/CD15+/HLA-DRneg) and monocytic subtypes (CD14+/CD15neg/HLA-DRneg). In the present study, we found a population of monocytes expressing the granulocyte marker CD15 that significantly increased in both peripheral blood (PB) and tumoral tissues of patients with colorectal cancer (CRC). Further phenotypical analysis confirmed the granulocytic-like features of this monocyte subpopulation that is associated with an increase in granulocyte-monocyte precursors (GMPs) in the PB of these patients (pts). Mechanistically, this granulocyte-like monocyte population suppressed NK cell activity by inducing TIGIT and engaging NKp30. Accordingly, an increased frequency of TIGIT+ NK cells with impaired functions was found in both the PB and tumoral tissue of CRC pts. Collectively, we provided new mechanistic explanations for tumor immune escape occurring in CRC by showing the increase in this new kind of MDSC, in both PB and CRC tissue, which is able to significantly impair the effector functions of NK cells, thereby representing a potential therapeutic target for cancer immunotherapy.
Keywords: CRC; MDSCs; NK cells; TIGIT; human; monocytes; neutrophil-like cells.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


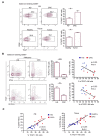
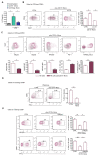

References
-
- Corzo C.A., Cotter M.J., Cheng P., Cheng F., Kusmartsev S., Sotomayor E., Padhya T., McCaffrey T.V., McCaffrey J.C., Gabrilovich D.I. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J. Immunol. 2009;182:5693–5701. doi: 10.4049/jimmunol.0900092. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

