Interplay of chromatin organization and mechanics of the cell nucleus
- PMID: 39126157
- PMCID: PMC11480768
- DOI: 10.1016/j.bpj.2024.08.003
Interplay of chromatin organization and mechanics of the cell nucleus
Abstract
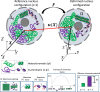
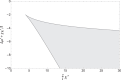
The nucleus of eukaryotic cells is constantly subjected to different kinds of mechanical stimuli, which can impact the organization of chromatin and, subsequently, the expression of genetic information. Experiments from different groups showed that nuclear deformation can lead to transient or permanent condensation or decondensation of chromatin and the mechanical activation of genes, thus altering the transcription of proteins. Changes in chromatin organization, in turn, change the mechanical properties of the nucleus, possibly leading to an auxetic behavior. Here, we model the mechanics of the nucleus as a chemically active polymer gel in which the chromatin can exist in two states: a self-attractive state representing the heterochromatin and a repulsive state representing euchromatin. The model predicts reversible or irreversible changes in chromatin condensation levels upon external deformations of the nucleus. We find an auxetic response for a broad range of parameters under small and large deformations. These results agree with experimental observations and highlight the key role of chromatin organization in the mechanical response of the nucleus.
Copyright © 2024 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

