Efficient Enrichment of Synchronized Mouse Spermatocytes Suitable for Genome-Wide Analysis
- PMID: 39126467
- PMCID: PMC12536511
- DOI: 10.1007/978-1-0716-3906-1_4
Efficient Enrichment of Synchronized Mouse Spermatocytes Suitable for Genome-Wide Analysis
Abstract
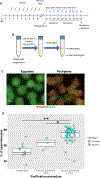
Chromatin undergoes extensive remodeling during meiosis, leading to specific patterns of gene expression and chromosome organization, which ultimately controls fundamental meiotic processes such as recombination and homologous chromosome associations. Recent game-changing advances have been made by analysis of chromatin binding sites of meiotic specific proteins genome-wide in mouse spermatocytes. However, further progress is still highly dependent on the reliable isolation of sufficient quantities of spermatocytes at specific stages of prophase I. Here, we describe a combination of methodologies we adapted for rapid and reliable isolation of synchronized fixed mouse spermatocytes. We show that chromatin isolated from these cells can be used to study chromatin-binding sites by ChIP-seq. High-quality data we obtained from INO80 ChIP-seq in zygotene cells was used for functional analysis of chromatin-binding sites.
Keywords: ChIP-seq; Genomic analysis; Mouse spermatocytes; Spermatocyte fraction enrichment.
© 2024. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Figures

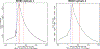
References
-
- Barchi M, Geremia R, Magliozzi R, Bianchi E, Isolation and analyses of enriched populations of male mouse germ cells by sedimentation velocity: the centrifugal elutriation. Methods in molecular biology 558, 299–321 (2009). - PubMed
-
- Romer KA, de Rooij DG, Kojima ML, Page DC, Isolating mitotic and meiotic germ cells from male mice by developmental synchronization, staging, and sorting. Dev Biol 443, 19–34 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

