Functional genomic analysis of genes important for Candida albicans fitness in diverse environmental conditions
- PMID: 39126650
- PMCID: PMC11416860
- DOI: 10.1016/j.celrep.2024.114601
Functional genomic analysis of genes important for Candida albicans fitness in diverse environmental conditions
Abstract
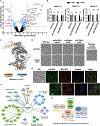
Fungal pathogens such as Candida albicans pose a significant threat to human health with limited treatment options available. One strategy to expand the therapeutic target space is to identify genes important for pathogen growth in host-relevant environments. Here, we leverage a pooled functional genomic screening strategy to identify genes important for fitness of C. albicans in diverse conditions. We identify an essential gene with no known Saccharomyces cerevisiae homolog, C1_09670C, and demonstrate that it encodes subunit 3 of replication factor A (Rfa3). Furthermore, we apply computational analyses to identify functionally coherent gene clusters and predict gene function. Through this approach, we predict the cell-cycle-associated function of C3_06880W, a previously uncharacterized gene required for fitness specifically at elevated temperatures, and follow-up assays confirm that C3_06880W encodes Iml3, a component of the C. albicans kinetochore with roles in virulence in vivo. Overall, this work reveals insights into the vulnerabilities of C. albicans.
Keywords: CP: Genomics; CP: Microbiology; Candida albicans; DNA damage repair; Galleria mellonella; fitness; functional genomics; fungal pathogen; kinetochore; virulence.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.E.C. is a co-founder and shareholder in Bright Angel Therapeutics, a platform company for the development of novel antifungal therapeutics. L.E.C. is a Science Advisor for Kapoose Creek, a company that harnesses the therapeutic potential of fungi.
Figures




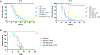
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

