Exposure to bacterial PAMPs before RSV infection exacerbates innate inflammation and disease via IL-1α and TNF-α
- PMID: 39127259
- PMCID: PMC11631774
- DOI: 10.1016/j.mucimm.2024.08.002
Exposure to bacterial PAMPs before RSV infection exacerbates innate inflammation and disease via IL-1α and TNF-α
Abstract
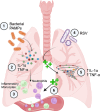
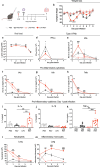
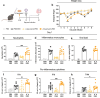
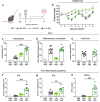
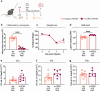
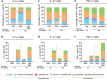
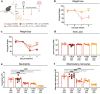
Respiratory syncytial virus (RSV) can cause severe lower respiratory tract infections. Understanding why some individuals get more serious disease may help with diagnosis and treatment. One possible risk factor underlying severe disease is bacterial exposure before RSV infection. Bacterial exposure has been associated with increased respiratory viral-induced disease severity but the mechanism remains unknown. Respiratory bacterial infections or exposure to their pathogen associated molecular patterns (PAMPs) trigger innate immune inflammation, characterised by neutrophil and inflammatory monocyte recruitment and the production of inflammatory cytokines. We hypothesise that these changes to the lung environment alter the immune response and disease severity during subsequent RSV infection. To test this, we intranasally exposed mice to LPS, LTA or Acinetobacter baumannii (an airway bacterial pathogen) before RSV infection and observed an early induction of disease, measured by weight loss, at days 1-3 after infection. Neutrophils or inflammatory monocytes were not responsible for driving this exacerbated weight loss. Instead, exacerbated disease was associated with increased IL-1α and TNF-α, which orchestrated the recruitment of innate immune cells into the lung. This study shows that exposure to bacterial PAMPs prior to RSV infection increases the expression of IL-1α and TNF-α, which dysregulate the immune response resulting in exacerbated disease.
Keywords: Bacteria; Innate immunity; Pro-inflammatory cytokines; Respiratory infections; Virus infections.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 2018;18:1191–210. doi: 10.1016/S1473-3099(18)30310-4. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

