Effective knockdown-replace gene therapy in a novel mouse model of DNM1 developmental and epileptic encephalopathy
- PMID: 39127888
- PMCID: PMC11489538
- DOI: 10.1016/j.ymthe.2024.08.009
Effective knockdown-replace gene therapy in a novel mouse model of DNM1 developmental and epileptic encephalopathy
Abstract
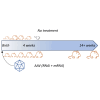
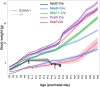
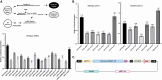
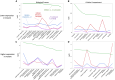
Effective gene therapy for gain-of-function or dominant-negative disease mutations may require eliminating expression of the mutant copy together with wild-type replacement. We evaluated such a knockdown-replace strategy in a mouse model of DNM1 disease, a debilitating and intractable neurodevelopmental epilepsy. To challenge the approach robustly, we expressed a patient-based variant in GABAergic neurons-which resulted in growth delay and lethal seizures evident by postnatal week three-and delivered to newborn pups an AAV9-based vector encoding a ubiquitously expressed, Dnm1-specific interfering RNA (RNAi) bivalently in tail-to-tail configuration with a neuron-specific, RNAi-resistant, codon-optimized Dnm1 cDNA. Pups receiving RNAi or cDNA alone fared no better than untreated pups, whereas the vast majority of mutants receiving modest doses survived with almost full growth recovery. Synaptic recordings of cortical neurons derived from treated pups revealed that significant alterations in transmission from inhibitory to excitatory neurons were rectified by bivalent vector application. To examine the mutant transcriptome and impact of treatment, we used RNA sequencing and functional annotation clustering. Mutants displayed abnormal expression of more than 1,000 genes in highly significant and relevant functional clusters, clusters that were abrogated by treatment. Together these results suggest knockdown-replace as a potentially effective strategy for treating DNM1 and related genetic neurodevelopmental disease.
Keywords: deno-associated virus; developmental and epileptic encephalopathy; dynamin-1; epilepsy; neurodevelopment.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A patent application was submitted (Appl. No. 63/639,576: Products and methods to inhibit expression of dynamin-1 variants and replace dynamin-1). Inventors: S.Q.H., N.T., and W.N.F.
Figures





References
-
- Dhindsa R.S., Bradrick S.S., Yao X., Heinzen E.L., Petrovski S., Krueger B.J., Johnson M.R., Frankel W.N., Petrou S., Boumil R.M., Goldstein D.B. Epileptic encephalopathy-causing mutations in DNM1 impair synaptic vesicle endocytosis. Neurol. Genet. 2015;1:e4. doi: 10.1212/01.NXG.0000464295.65736.da. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

