Mutant TP53 switches therapeutic vulnerability during gastric cancer progression within interleukin-6 family cytokines
- PMID: 39128004
- PMCID: PMC11372443
- DOI: 10.1016/j.celrep.2024.114616
Mutant TP53 switches therapeutic vulnerability during gastric cancer progression within interleukin-6 family cytokines
Abstract
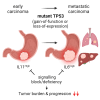
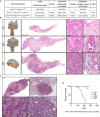
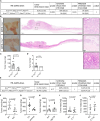
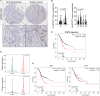
Although aberrant activation of the KRAS and PI3K pathway alongside TP53 mutations account for frequent aberrations in human gastric cancers, neither the sequence nor the individual contributions of these mutations have been clarified. Here, we establish an allelic series of mice to afford conditional expression in the glandular epithelium of KrasG12D;Pik3caH1047R or Trp53R172H and/or ablation of Pten or Trp53. We find that KrasG12D;Pik3caH1047R is sufficient to induce adenomas and that lesions progress to carcinoma when also harboring Pten deletions. An additional challenge with either Trp53 loss- or gain-of-function alleles further accelerated tumor progression and triggered metastatic disease. While tumor-intrinsic STAT3 signaling in response to gp130 family cytokines remained as a gatekeeper for all stages of tumor development, metastatic progression required a mutant Trp53-induced interleukin (IL)-11 to IL-6 dependency switch. Consistent with the poorer survival of patients with high IL-6 expression, we identify IL-6/STAT3 signaling as a therapeutic vulnerability for TP53-mutant gastric cancer.
Keywords: CP: Cancer; STAT3; TP53; WNT; animal model; cytokine signaling; gastric carcinoma; interleukin-11; interleukin-6; metastasis; organoids.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.E. serves on the scientific advisory board of Lassen Therapeutics, which develops anti-IL-11 therapeutics.
Figures


References
-
- Arnold M., Rutherford M., Lam F., Bray F., Ervik M., Soerjomataram I. 2019. ICBP SURVMARK-2 Online Tool: International Cancer Survival Benchmarking. Lyon, France.https://gco.iarc.fr/survival/survmark International Agency for Research on Cancer.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

