Inter-strain variability in responses to a single administration of the cannabidiol-rich cannabis extract in mice
- PMID: 39128689
- PMCID: PMC11381146
- DOI: 10.1016/j.fct.2024.114909
Inter-strain variability in responses to a single administration of the cannabidiol-rich cannabis extract in mice
Abstract
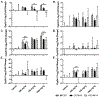
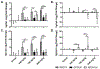
Cannabidiol (CBD) has gained widespread popularity; however, its pharmacological and toxicological profiles in the context of human genetic diversity remain largely unexplored. Here, we investigated the variability in metabolism and toxicity of CBD-rich cannabis extract (CRCE) in genetically diverse mouse models: C57BL/6J, B6C3F1/J, and NZO/HlLtJ strains. Mice received a single dose of CRCE containing 57.9% CBD at dosages of 0, 246, 738, and 2460 mg/kg of CBD. At 24 h after treatment, no appreciable histomorphological changes were detected in the liver. Plasma bilirubin levels increased markedly in all strains at the highest CBD dose. Mice in all treatment groups displayed significant but distinct increases in ALT and AST levels. While B6C3F1/J and NZO/HlLtJ mice had negligible plasma CBD levels at 738 mg/kg, C57BL/6J mice exhibited levels exceeding 7000 ng/mL. At 2460 mg/kg, high CBD concentrations were found in B6C3F1/J and C57BL/6J mice, but markedly lower levels were seen in NZO/HlLtJ mice. Gene expression profiling showed significant increases in Cyp2b10 across all strains but varying responses in Cyp1a1 expression, indicating strain-specific CYP dysregulation. Genetically diverse mice exhibited differential pharmacological and toxicological responses to CRCE, suggesting a high potential for inter-individual variability in the pharmacology and toxicology of CBD in humans.
Keywords: Cannabidiol-rich cannabis extract; Cytochromes; Genetic diversity; Hepatotoxicity; Metabolism; Mouse model.
Copyright © 2024 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


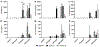
References
-
- Alugupalli KR, Kothari S, Cravens MP, Walker JA, Dougharty DT, Dickinson GS, Gatto LA, Bäumler AJ, Wangdi T, Miller DR, Pardo-Manuel de Villena F, Siracusa LD. Identification of collaborative cross mouse strains permissive to Salmonella enterica serovar Typhi infection. Sci Rep. 2023. Jan 9;13(1):393. doi: 10.1038/s41598-023-27400-1. - DOI - PMC - PubMed
-
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, de Villena FP, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011. Aug;21(8):1213–22. doi: 10.1101/gr.111310.110. Epub 2011 Mar 15.. - DOI - PMC - PubMed
-
- Chan P, Mahler J. NTP technical report on the toxicity studies of Glyphosate (CAS No. 1071-83-6) Administered In Dosed Feed To F344/N Rats And B6C3F1 Mice. Toxic Rep Ser. 1992. Jul;16:1–D3. - PubMed
-
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008. Jun;19(6):382–9. doi: 10.1007/s00335-008-9135-8. Epub 2008 Aug 21. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

