MiRNA Dysregulation in Brain Injury: An In Silico Study to Clarify the Role of a MiRNA Set
- PMID: 39129166
- PMCID: PMC11793054
- DOI: 10.2174/1570159X22666240808124427
MiRNA Dysregulation in Brain Injury: An In Silico Study to Clarify the Role of a MiRNA Set
Abstract
Background: The identification of specific circulating miRNAs has been proposed as a valuable tool for elucidating the pathophysiology of brain damage or injury and predicting patient outcomes.
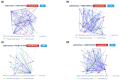
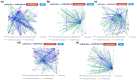
Objective: This study aims to apply several bioinformatic tools in order to clarify miRNA interactions with potential genes involved in brain injury, emphasizing the need of using a computational approach to determine the most likely correlations between miRNAs and target genes. Specifically, this study centers on elucidating the roles of miR-34b, miR-34c, miR-135a, miR-200c, and miR-451a.
Methods: After a careful evaluation of different software available (analyzing the strengths and limitations), we applied three tools, one to perform an analysis of the validated targets (miRTarBase), and two to evaluate functional annotations (miRBase and TAM 2.0).
Results: Research findings indicate elevated levels of miR-135a and miR-34b in patients with traumatic brain injury (TBI) within the first day post-injury, while miR-200c and miR-34c were found to be upregulated after 7 days. Moreover, miR-451a and miR-135a were found overexpressed in the serum, while miRNAs 34b, 34c, and 200c, had lower serum levels at baseline post brain injury.
Conclusion: This study emphasizes the use of computational methods in determining the most likely relationships between miRNAs and target genes by investigating several bioinformatic techniques to elucidate miRNA interactions with potential genes. Specifically, this study focuses on the functions of miR-34b, miR-34c, miR-135a, miR-200c, and miR-451a, providing an up-to-date overview and suggesting future research directions for identifying theranomiRNAs related to brain injury, both at the tissue and serum levels.
Keywords: Brain injury; biomarkers; diagnosis; miRNA; prognosis; theranomiRNA..
Copyright© Bentham Science Publishers; For any queries, please email at epub@benthamscience.net.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures

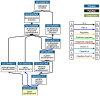
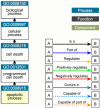

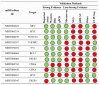

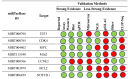





References
-
- Sessa F., Maglietta F., Bertozzi G., Salerno M., Di Mizio G., Messina G., Montana A., Ricci P., Pomara C. Human brain injury and mirnas: An experimental study. Int. J. Mol. Sci. 2019;20(7):1546. doi: 10.3390/ijms20071546. https://www.scopus.com/inward/record.uri?eid=2-s2.0-85064195391&doi=10.3... - DOI - PMC - PubMed
-
- Carvalho L.B., dos Santos Sanna P.L., dos Santos Afonso C.C., Bondan E.F., da Silva Feltran G., Ferreira M.R., Birbrair A., Andia D.C., Latini A., Foganholi da Silva R.A. MicroRNA biogenesis machinery activation and lncRNA and REST overexpression as neuroprotective responses to fight inflammation in the hippocampus. J. Neuroimmunol. 2023;382:578149. doi: 10.1016/j.jneuroim.2023.578149. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

