CRISPR/Cas13a-assisted amplification-free miRNA biosensor via dark-field imaging and magnetic gold nanoparticles
- PMID: 39129860
- PMCID: PMC11308380
- DOI: 10.1039/d4sd00081a
CRISPR/Cas13a-assisted amplification-free miRNA biosensor via dark-field imaging and magnetic gold nanoparticles
Abstract
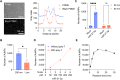
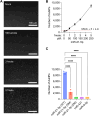
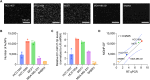
MicroRNAs (miRNAs) are short (about 18-24 nucleotides) non-coding RNAs and have emerged as potential biomarkers for various diseases, including cancers. Due to their short lengths, the specificity often becomes an issue in conventional amplification-based methods. Next-generation sequencing techniques could be an alternative, but the long analysis time and expensive costs make them less suitable for routine clinical diagnosis. Therefore, it is essential to develop a rapid, selective, and accurate miRNA detection assay using a simple, affordable system. In this work, we report a CRISPR/Cas13a-based miRNA biosensing using point-of-care dark-field (DF) imaging. We utilized magnetic-gold nanoparticle (MGNPs) complexes as signal probes, which consist of 200 nm-sized magnetic beads and 60 nm-sized gold nanoparticles (AuNPs) linked by DNA hybridization. Once the CRISPR/Cas13a system recognized the target miRNAs (miR-21-5p), the activated Cas13a cleaved the bridge linker containing RNA sequences, releasing 60 nm-AuNPs detected and quantified by a portable DF imaging system. The combination of CRISPR/Cas13a, MGNPs, and DF imaging demonstrated amplification-free detection of miR-21-5p within 30 min at a detection limit of 500 attomoles (25 pM) and with single-base specificity. The CRISPR/Cas13a-assisted MGNP-DF assay achieved rapid, selective, and accurate detection of miRNAs with simple equipment, thus providing a potential application for cancer diagnosis.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
H. I. is a scientific advisory board member of Nanopath and AITRICS and a consultant to Cellkey, Co. Ltd. H. I. receives research support from Canon Medical Research USA, Healcerion, and Noul. These activities have no relationship to the study presented here.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
