Development and Validation of a Machine Learning-Based Prediction Model for Detection of Biliary Atresia
- PMID: 39130111
- PMCID: PMC11307559
- DOI: 10.1016/j.gastha.2023.05.002
Development and Validation of a Machine Learning-Based Prediction Model for Detection of Biliary Atresia
Abstract
Background and aims: Biliary atresia is a rare and devastating bile duct disease that occurs during the neonatal period. Timely identification and prompt surgical intervention is critical for improving the outcome. The aim of the study was to develop a new machine learning-based prediction model for the detection of biliary atresia.
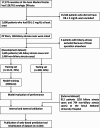
Methods: Neonates aged <100 days with cholestasis at least once were retrospectively screened in 2 tertiary referral hospitals between 2015 and 2020. Simple demographic data, routine laboratory indices, and imaging findings of ultrasonography and hepatobiliary scintigraphy were used as features in the multivariate analysis. The extreme gradient boosting (XGBoost) framework was used to develop prediction models according to the diagnostic steps.
Results: Among 1605 enrolled neonates with all-cause cholestasis, 145 (9%) were included as having biliary atresia. Direct bilirubin, gamma-glutamyl transpeptidase, abdominal sonography, and hepatobiliary scan were the most impactful features in prediction models. The Step II XGBoost model, consisting of nonimaging inputs, showed excellent discriminatory performance (area under the curve = 0.97). The Step III and IV XGBoost models showed near-perfect performances (area under the curve = 0.998 and 0.999, respectively). In external validation (n = 912 with 118 [12.9%] biliary atresia), XGBoost-based prediction models consistently showed acceptable performances. Utilizing shapley additive explanation values also provided visualized insight and explanation of the contribution of features in detecting biliary atresia. The models were integrated into a web-based diagnostic tool for case-level application.
Conclusion: We introduced a new machine learning-based prediction model for detecting biliary atresia in the largest cohorts of neonatal cholestasis.
Keywords: Biliary Atresia; Machine Learning; Neonatal Cholestasis; Prediction; XGBoost.
© 2023 The Authors.
Figures



References
-
- Hartley J.L., Davenport M., Kelly D.A. Biliary atresia. Lancet. 2009;374(9702):1704–1713. - PubMed
-
- Fawaz R., Baumann U., Ekong U., et al. Guideline for the Evaluation of Cholestatic Jaundice in Infants: Joint Recommendations of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. J Pediatr Gastroenterol Nutr. 2017;64(1):154–168. - PubMed
-
- El-Guindi M.A., Sira M.M., Sira A.M., et al. Design and validation of a diagnostic score for biliary atresia. J Hepatol. 2014;61(1):116–123. - PubMed
LinkOut - more resources
Full Text Sources

