Identification and experimental validation of programmed cell death- and mitochondria-associated biomarkers in osteoporosis and immune microenvironment
- PMID: 39130750
- PMCID: PMC11310001
- DOI: 10.3389/fgene.2024.1439171
Identification and experimental validation of programmed cell death- and mitochondria-associated biomarkers in osteoporosis and immune microenvironment
Abstract
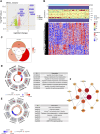
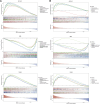
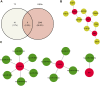
Background: Prior research has demonstrated that programmed cell death (PCD) and mitochondria assume pivotal roles in controlling cellular metabolism and maintaining bone cell equilibrium. Nonetheless, the comprehensive elucidation of their mode of operation in osteoporosis (OP) warrants further investigation. Therefore, this study aimed at analyzing the role of genes associated with PCD (PCD-RGs) and mitochondria (mortality factor-related genes; MRGs) in OP. Methods: Differentially expressed genes (DEGs) were identified by subjecting the GSE56815 dataset obtained from the Gene Expression Omnibus database to differential expression analysis and comparing OP patients with healthy individuals. The genes of interest were ascertained through the intersection of DEGs, MRGs, and PCD-RGs; these genes were filtered using machine learning methodologies to discover potential biomarkers. The prospective biomarkers displaying uniform patterns and statistically meaningful variances were identified by evaluating their levels in the GSE56815 dataset and conducting quantitative real-time polymerase chain reaction-based assessments. Moreover, the functional mechanisms of these biomarkers were further delineated by constructing a nomogram, which conducted gene set enrichment analysis, explored immune infiltration, generated regulatory networks, predicted drug responses, and performed molecular docking analyses. Results: Eighteen candidate genes were documented contingent upon the intersection between 2,354 DEGs, 1,136 MRGs, and 1,548 PCD-RGs. The biomarkers DAP3, BIK, and ACAA2 were upregulated in OP and were linked to oxidative phosphorylation. Furthermore, the predictive ability of the nomogram designed based on the OP biomarkers exhibited a certain degree of accuracy. Correlation analysis revealed a strong positive correlation between CD56dim natural killer cells and ACAA2 and a significant negative correlation between central memory CD4+ T cells and DAP3. DAP3, BIK, and ACAA2 were regulated by multiple factors; specifically, SETDB1 and ZNF281 modulated ACAA2 and DAP3, whereas TP63 and TFAP2C governed DAP3 and BIK. Additionally, a stable binding force was observed between the drugs (estradiol, valproic acid, and CGP52608) and the biomarkers. Conclusion: This investigation evidenced that the biomarkers DAP3, BIK, and ACAA2 are associated with PCD and mitochondria in OP, potentially facilitate the diagnosis of OP in clinical settings.
Keywords: machine learning; mitochondria; nomogram; osteoporosis; programmed cell death.
Copyright © 2024 Yang, Zhang, Lu, Chen, Wang, Lin, Chen, Chen and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

