Prediction of TERT mutation status in gliomas using conventional MRI radiogenomic features
- PMID: 39131044
- PMCID: PMC11310134
- DOI: 10.3389/fneur.2024.1439598
Prediction of TERT mutation status in gliomas using conventional MRI radiogenomic features
Abstract
Objective: Telomerase reverse transcriptase (TERT) promoter mutation status in gliomas is a key determinant of treatment strategy and prognosis. This study aimed to analyze the radiogenomic features and construct radiogenomic models utilizing medical imaging techniques to predict the TERT promoter mutation status in gliomas.
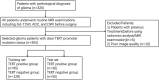
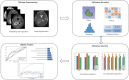
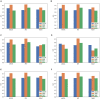
Methods: This was a retrospective study of 304 patients with gliomas. T1-weighted contrast-enhanced, apparent diffusion coefficient, and diffusion-weighted imaging MRI sequences were used for radiomic feature extraction. A total of 3,948 features were extracted from MRI images using the FAE software. These included 14 shape features, 18 histogram features, 24 gray level run length matrix, 14 gray level dependence matrix, 16 gray level run length matrix, 16 gray level size zone matrix (GLSZM), 5 neighboring gray tone difference matrix, and 744 wavelet transforms. The dataset was randomly divided into training and testing sets in a ratio of 7:3. Three feature selection methods and six classification algorithms were used to model the selected features. Predictive performance was evaluated using receiver operating characteristic curve analysis.
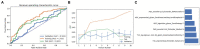
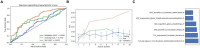
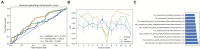
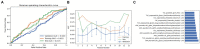
Results: Among the evaluated classification algorithms, the combination model of recursive feature elimination (RFE) with linear regression (LR) using six features showed the best diagnostic performance (area under the curve: 0.733, 0.562, and 0.633 in the training, validation, and testing sets, respectively). The next best-performing models were naive Bayes, linear discriminant analysis, autoencoder, and support vector machine. Regarding the three feature selection algorithms, RFE showed the most consistent performance, followed by relief and ANOVA. T1-enhanced entropy and GLSZM derived from T1-enhanced images were identified as the most critical radiomics features for distinguishing TERT promoter mutation status.
Conclusion: The LR and LRLasso models, mainly based on T1-enhanced entropy and GLSZM, showed good predictive ability for TERT promoter mutations in gliomas using radiomics models.
Keywords: TERT promoter mutation; glioma; machine learning; magnetic resonance imaging; radiomics.
Copyright © 2024 Tang, Chen, Xu, Huang and Zeng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
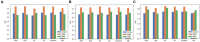
References
LinkOut - more resources
Full Text Sources

