DNA Origami-Engineered Plasmonic Nanoprobes for Targeted Cancer Imaging
- PMID: 39131199
- PMCID: PMC11309351
- DOI: 10.1002/adfm.202309929
DNA Origami-Engineered Plasmonic Nanoprobes for Targeted Cancer Imaging
Abstract
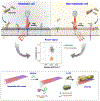
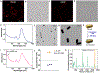
Plasmonic nanomaterials bearing targeting ligands are of great interest for surface-enhanced Raman scattering (SERS)-based bioimaging applications. However, the practical utility of SERS-based imaging strategies has been hindered by the lack of a straightforward method to synthesize highly sensitive SERS-active nanostructures with high yield and efficiency. In this work, leveraging DNA origami principles, we report the first-in-class design of a SERS-based plasmonically coupled nanoprobe for targeted cancer imaging (SPECTRA). The nanoprobe harnesses a cancer cell targeting DNA aptamer sequence and vibrational tag with stretching frequency in the cell-silent Raman window. Through the integration of aptamer sequence specific for DU145 cells, we show the unique capabilities of SPECTRA for targeted imaging of DU145 cells. Our results demonstrate that the scalability, cost-effectiveness, and reproducibility of this method of fabrication of SERS nanoprobes can serve as a versatile platform for creating nanoprobes with broad applications in the fields of cancer biology and biomedical imaging.
Keywords: DNA origami; plasmonic nanoantenna; surface-enhanced Raman scattering (SERS); targeted imaging.
Conflict of interest statement
Conflict of interest The authors declare no conflict of interest.
Figures





References
-
- Cheng J, Xie XS, Science 2015, 350(6264), aaa8870. - PubMed
-
- Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, Bernstein L, Guiot M, Petrecca K, Leblond F, Sci. Transl. Med 2015, 7(274), 274ra19. - PubMed
-
- Tanwar S, Haldar K, Sen T, J. Am. Chem. Soc 2017,139(48), 17639–17648. - PubMed
-
- Tanwar S, Kaur V, Kaur G, Sen T, J. Phys. Chem. Lett 2021, 12(33), 8141–8150. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
