This is a preprint.
Mechanism of neurodegeneration mediated by clonal inflammatory microglia
- PMID: 39131366
- PMCID: PMC11312538
- DOI: 10.1101/2024.07.30.605867
Mechanism of neurodegeneration mediated by clonal inflammatory microglia
Abstract
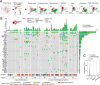
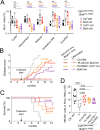
Langerhans cell Histiocytosis (LCH) and Erdheim-Chester disease (ECD) are clonal myeloid disorders, associated with MAP-Kinase activating mutations and an increased risk of neurodegeneration. Surprisingly, we found pervasive PU.1+ microglia mutant clones across the brain of LCH and ECD patients with and without neurological symptoms, associated with microgliosis, reactive astrocytosis, and neuronal loss. The disease predominated in the grey nuclei of the rhombencephalon, a topography attributable to a local proliferative advantage of mutant microglia. Presence of clinical symptoms was associated with a longer evolution of the disease and a larger size of PU.1+ clones (p= 0.0003). Genetic lineage tracing of PU.1+ clones suggest a resident macrophage lineage or a bone marrow precursor origin depending on patients. Finally, a CSF1R-inhibitor depleted mutant microglia and limited neuronal loss in mice suggesting an alternative to MAPK inhibitors. These studies characterize a progressive neurodegenerative disease, caused by clonal proliferation of inflammatory microglia (CPIM), with a decade(s)-long preclinical stage of incipient disease that represent a therapeutic window for prevention of neuronal death.
Conflict of interest statement
Conflict of Interest. FG has performed consulting for Third Rock venture in the past. Targeted Sequencing was funded in part by a grant from Third Rock venture. FG and RV are inventors in MSKCC’s United States application or PCT international application number PCT/US2018/047964 filed on 8/24/2018 (KINASE MUTATION-ASSOCIATED NEURODEGENERATIVE DISORDERS)
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
