Exploring the Potential of Machine Learning Algorithms to Improve Diffusion Nuclear Magnetic Resonance Imaging Models Analysis
- PMID: 39131437
- PMCID: PMC11309135
- DOI: 10.4103/jmp.jmp_10_24
Exploring the Potential of Machine Learning Algorithms to Improve Diffusion Nuclear Magnetic Resonance Imaging Models Analysis
Abstract
Purpose: This paper explores different machine learning (ML) algorithms for analyzing diffusion nuclear magnetic resonance imaging (dMRI) models when analytical fitting shows restrictions. It reviews various ML techniques for dMRI analysis and evaluates their performance on different b-values range datasets, comparing them with analytical methods.
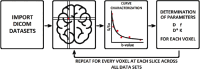
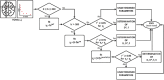
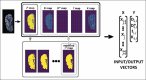
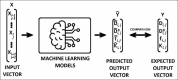
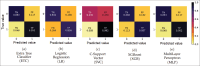
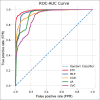
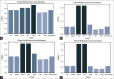
Materials and methods: After standard fitting for reference, four sets of diffusion-weighted nuclear magnetic resonance images were used to train/test various ML algorithms for prediction of diffusion coefficient (D), pseudo-diffusion coefficient (D*), perfusion fraction (f), and kurtosis (K). ML classification algorithms, including extra-tree classifier (ETC), logistic regression, C-support vector, extra-gradient boost, and multilayer perceptron (MLP), were used to determine the existence of diffusion parameters (D, D*, f, and K) within single voxels. Regression algorithms, including linear regression, polynomial regression, ridge, lasso, random forest (RF), elastic-net, and support-vector machines, were used to estimate the value of the diffusion parameters. Performance was evaluated using accuracy (ACC), area under the curve (AUC) tests, and cross-validation root mean square error (RMSECV). Computational timing was also assessed.
Results: ETC and MLP were the best classifiers, with 94.1% and 91.7%, respectively, for the ACC test and 98.7% and 96.3% for the AUC test. For parameter estimation, RF algorithm yielded the most accurate results The RMSECV percentages were: 8.39% for D, 3.57% for D*, 4.52% for f, and 3.53% for K. After the training phase, the ML methods demonstrated a substantial decrease in computational time, being approximately 232 times faster than the conventional methods.
Conclusions: The findings suggest that ML algorithms can enhance the efficiency of dMRI model analysis and offer new perspectives on the microstructural and functional organization of biological tissues. This paper also discusses the limitations and future directions of ML-based dMRI analysis.
Keywords: Diffusion magnetic resonance imaging; intravoxel incoherent motion; kurtosis; machine learning.
Copyright: © 2024 Journal of Medical Physics.
Conflict of interest statement
There are no conflicts of interest.
Figures
References
-
- Le Bihan D. What can we see with IVIM MRI? Neuroimage. 2019;187:56–67. - PubMed
-
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval Jeantet M. MR imaging of intravoxel incoherent motions: Application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161:401–7. - PubMed
-
- Rosenkrantz AB, Padhani AR, Chenevert TL, Koh DM, De Keyzer F, Taouli B, et al. Body diffusion kurtosis imaging: Basic principles, applications, and considerations for clinical practice. J Magn Reson Imaging. 2015;42:1190–202. - PubMed
-
- Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K. Diffusional kurtosis imaging: The quantification of non-Gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med. 2005;53:1432–40. - PubMed
LinkOut - more resources
Full Text Sources
