High-quality chromosome scale genome assemblies of two important Sorghum inbred lines, Tx2783 and RTx436
- PMID: 39131819
- PMCID: PMC11310780
- DOI: 10.1093/nargab/lqae097
High-quality chromosome scale genome assemblies of two important Sorghum inbred lines, Tx2783 and RTx436
Abstract
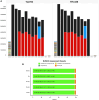
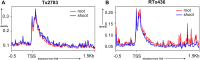
Sorghum bicolor (L.) Moench is a significant grass crop globally, known for its genetic diversity. High quality genome sequences are needed to capture the diversity. We constructed high-quality, chromosome-level genome assemblies for two vital sorghum inbred lines, Tx2783 and RTx436. Through advanced single-molecule techniques, long-read sequencing and optical maps, we improved average sequence continuity 19-fold and 11-fold higher compared to existing Btx623 v3.0 reference genome and obtained 19 and 18 scaffolds (N50 of 25.6 and 14.4) for Tx2783 and RTx436, respectively. Our gene annotation efforts resulted in 29 612 protein-coding genes for the Tx2783 genome and 29 265 protein-coding genes for the RTx436 genome. Comparative analyses with 26 plant genomes which included 18 sorghum genomes and 8 outgroup species identified around 31 210 protein-coding gene families, with about 13 956 specific to sorghum. Using representative models from gene trees across the 18 sorghum genomes, a total of 72 579 pan-genes were identified, with 14% core, 60% softcore and 26% shell genes. We identified 99 genes in Tx2783 and 107 genes in RTx436 that showed functional enrichment specifically in binding and metabolic processes, as revealed by the GO enrichment Pearson Chi-Square test. We detected 36 potential large inversions in the comparison between the BTx623 Bionano map and the BTx623 v3.1 reference sequence. Strikingly, these inversions were notably absent when comparing Tx2783 or RTx436 with the BTx623 Bionano map. These inversion were mostly in the pericentromeric region which is known to have low complexity regions and harder to assemble and suggests the presence of potential artifacts in the public BTx623 reference assembly. Furthermore, in comparison to Tx2783, RTx436 exhibited 324 883 additional Single Nucleotide Polymorphisms (SNPs) and 16 506 more Insertions/Deletions (INDELs) when using BTx623 as the reference genome. We also characterized approximately 348 nucleotide-binding leucine-rich repeat (NLR) disease resistance genes in the two genomes. These high-quality genomes serve as valuable resources for discovering agronomic traits and structural variation studies.
Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics 2024.
Figures




References
-
- Ordonio R., Ito Y., Morinaka Y., Sazuka T., Matsuoka M.. Molecular breeding of sorghum bicolor, a novel energy crop. Int. Rev. Cell Mol. Biol. 2016; 321:221–257. - PubMed
-
- Mbulwe L., Peterson G.C., Scott-Armstrong J., Rooney W.L.. Registration of Sorghum germplasm Tx3408 and Tx3409 with tolerance to sugarcane aphid [Melanaphis sacchari (Zehntner)]. Jo. Plant Registrations. 2016; 10:51–56.
LinkOut - more resources
Full Text Sources

