MIP-4 is Induced by Bleomycin and Stimulates Cell Migration Partially via Nir-1 Receptor
- PMID: 39132322
- PMCID: PMC11315970
- DOI: 10.1155/2024/5527895
MIP-4 is Induced by Bleomycin and Stimulates Cell Migration Partially via Nir-1 Receptor
Abstract
Background: CC-chemokine ligand 18 also known as MIP-4 is a chemokine with roles in inflammation and immune responses. It has been shown that MIP-4 is involved in the development of several diseases including lung fibrosis and cancer. How exactly MIP-4 is regulated and exerts its role in lung fibrosis remains unclear. Therefore, in the present study, we examined how MIP-4 is regulated and whether it acts via its potential receptor Nir-1.
Materials and methods: A549 cells were grown and maintained in DMEM : F12 (1 : 1) and supplemented with 10% FBS and 1000 U of penicillin/streptomycin and maintained as recommended by the manufacturer (ATCC). Cell migration and invasion, immunohistochemistry (IHC), Western blot, qPCR, and siRNA Nir-1 were used to determine MIP-4 regulation and its role in cell migration.
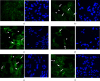
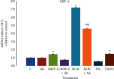
Results: Cell migration was increased following stimulation of cells with recombinant (r) MIP-4 and bleomycin (BLM), whereas quenching rMIP-4 with its antibody (Ab) or addition of the Ab to BLM or H2O2 diminished rMIP-4-induced cell migration. Along with cell migration, rMIP-4, BLM, and H2O2 induced the formation of actin filaments dynamic structures whereas costimulation with MIP-4 Ab limited BLM- and H2O2-induced effects. MIP-4 mRNA and protein were increased by BLM and H2O2, and the addition of its Ab significantly reduced treatments effect. Experiments with siRNA investigating whether Nir-1 is a potential MIR-4 receptor indicated that the inhibition of Nir-1 decreased cell migration/invasion but did not totally inhibit rMIP-4-induced cell migration.
Conclusion: Therefore, our data indicate that MIP-4 is regulated by BLM and H2O2 and costimulation with its Ab limits the effects on MIP-4 and that the Nir-1 receptor partially mediates MIP-4's effects on increased cell migration. These data also evidenced that MIP-4 is regulated by fibrotic and oxidative stimuli and that quenching MIP-4 with its Ab or therapeutically targeting the Nir-1 receptor may partially limit MIP-4 effects under fibrotic or oxidative stimulation.
Copyright © 2024 M. Pacurari et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest. The funding sponsors have no role in the design of the study including the collection and interpretation of data, writing of the manuscript, and the decision in the publication of the present manuscript.
Figures




References
-
- Schraufstatter I., Takamori H., Sikora L., Sriramarao P., DiScipio R. G. Eosinophils and monocytes produce pulmonary and activation-regulated chemokine, which activates cultured monocytes/macrophages. American Journal of Physiology-Lung Cellular and Molecular Physiology . 2004;286(3):L494–L501. doi: 10.1152/ajplung.00323.2002. - DOI - PubMed
-
- Pivarcsi A., Gombert M., Dieu-Nosjean M. C., et al. CC chemokine ligand 18, an atopic dermatitis-associated and dendritic cell-derived chemokine, is regulated by staphylococcal products and allergen exposure. The Journal of Immunology . 2004;173(9):5810–5817. doi: 10.4049/jimmunol.173.9.5810. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

