This is a preprint.
Reemergence of Oropouche virus between 2023 and 2024 in Brazil
- PMID: 39132482
- PMCID: PMC11312653
- DOI: 10.1101/2024.07.27.24310296
Reemergence of Oropouche virus between 2023 and 2024 in Brazil
Update in
-
Re-emergence of Oropouche virus between 2023 and 2024 in Brazil: an observational epidemiological study.Lancet Infect Dis. 2025 Feb;25(2):166-175. doi: 10.1016/S1473-3099(24)00619-4. Epub 2024 Oct 16. Lancet Infect Dis. 2025. PMID: 39423838 Free PMC article.
Abstract
Background: Oropouche virus (OROV; species Orthobunyavirus oropoucheense) is an arthropod-borne virus that has caused outbreaks of Oropouche fever in Central and South America since the 1950s. This study investigates virological factors contributing to the reemergence of Oropouche fever in Brazil between 2023 and 2024.
Methods: In this study, we combined OROV genomic, molecular, and serological data from Brazil from 1 January 2015 to 29 June 2024, along with in vitro and in vivo characterization. Molecular screening data included 93 patients with febrile illness between January 2023 and February 2024 from the Amazonas State. Genomic data comprised two genomic OROV sequences from patients. Serological data were obtained from neutralizing antibody tests comparing the prototype OROV strain BeAn 19991 and the 2024 epidemic strain. Epidemiological data included aggregated cases reported to the Brazilian Ministry of Health from 1 January 2014 to 29 June 2024.
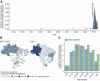
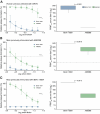
Findings: In 2024, autochthonous OROV infections were detected in previously non-endemic areas across all five Brazilian regions. Cases were reported in 19 of 27 federal units, with 83.2% (6,895 of 8,284) of infections in Northern Brazil and a nearly 200-fold increase in incidence compared to reported cases over the last decade. We detected OROV RNA in 10.8% (10 of 93) of patients with febrile illness between December 2023 and May 2024 in Amazonas. We demonstrate that the 2023-2024 epidemic was caused by a novel OROV reassortant that replicated approximately 100-fold higher titers in mammalian cells compared to the prototype strain. The 2023-2024 OROV reassortant displayed plaques earlier than the prototype, produced 1.7 times more plaques, and plaque sizes were 2.5 larger compared to the prototype. Furthermore, serum collected in 2016 from previously OROV-infected individuals showed at least a 32-fold reduction in neutralizing capacity against the reassortment strain compared to the prototype.
Interpretation: These findings provide a comprehensive assessment of Oropouche fever in Brazil and contribute to a better understanding of the 2023-2024 OROV reemergence. The recent increased incidence may be related to a higher replication efficiency of a new reassortant virus that also evades previous immunity.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Wesselmann KM, Postigo-Hidalgo I, Pezzi L, et al. Emergence of Oropouche fever in Latin America: a narrative review. Lancet Infect Dis 2024. - PubMed
-
- Pinheiro FP, Travassos da Rosa AP, Gomes ML, LeDuc JW, Hoch AL. Transmission of Oropouche virus from man to hamster by the midge Culicoides paraensis. Science 1982; 215(4537): 1251–3. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
