FUS-Mediated Inhibition of Myogenesis Elicited by Suppressing TNNT1 Production
- PMID: 39133076
- PMCID: PMC11376412
- DOI: 10.1080/10985549.2024.2383296
FUS-Mediated Inhibition of Myogenesis Elicited by Suppressing TNNT1 Production
Abstract
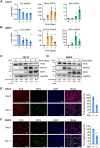
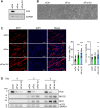
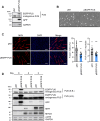
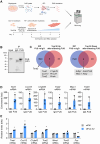
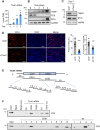
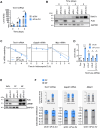
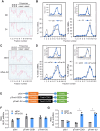
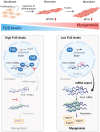
Myogenesis is a highly orchestrated process whereby muscle precursor cells, myoblasts, develop into muscle fibers to form skeletal muscle during embryogenesis and regenerate adult muscle. Here, we studied the RNA-binding protein FUS (fused in sarcoma), which has been implicated in muscular and neuromuscular pathologies but is poorly characterized in myogenesis. Given that FUS levels declined in human and mouse models of skeletal myogenesis, and that silencing FUS enhanced myogenesis, we hypothesized that FUS might be a repressor of myogenic differentiation. Interestingly, overexpression of FUS delayed myogenesis, accompanied by slower production of muscle differentiation markers. To identify the mechanisms through which FUS inhibits myogenesis, we uncovered RNA targets of FUS by ribonucleoprotein immunoprecipitation (RIP) followed by RNA-sequencing (RNA-seq) analysis. Stringent selection of the bound transcripts uncovered Tnnt1 mRNA, encoding troponin T1 (TNNT1), as a major effector of FUS influence on myogenesis. We found that in myoblasts, FUS retained Tnnt1 mRNA in the nucleus, preventing TNNT1 expression; however, reduction of FUS during myogenesis or by silencing FUS released Tnnt1 mRNA for export to the cytoplasm, enabling TNNT1 translation and promoting myogenesis. We propose that FUS inhibits myogenesis by suppressing TNNT1 expression through a mechanism of nuclear Tnnt1 mRNA retention.
Keywords: Myogenesis; TNNT1; ribonucleoprotein complex; translation.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
Dysregulated FOXO1 activity drives skeletal muscle intrinsic dysfunction in amyotrophic lateral sclerosis.Acta Neuropathol. 2024 Sep 16;148(1):43. doi: 10.1007/s00401-024-02794-y. Acta Neuropathol. 2024. PMID: 39283487 Free PMC article.
-
Pathogenic TNNT1 variants are associated with aberrant thin filament compliance and myofibre hyper-contractility.J Physiol. 2025 Jun;603(12):3533-3550. doi: 10.1113/JP288109. Epub 2025 May 5. J Physiol. 2025. PMID: 40320982 Free PMC article.
-
Function and Molecular Mechanism of Circhomer1 in Myogenesis.Int J Mol Sci. 2025 Jun 28;26(13):6264. doi: 10.3390/ijms26136264. Int J Mol Sci. 2025. PMID: 40650039 Free PMC article.
-
A systematic review of adult-onset clinically amyopathic dermatomyositis (dermatomyositis siné myositis): a missing link within the spectrum of the idiopathic inflammatory myopathies.J Am Acad Dermatol. 2006 Apr;54(4):597-613. doi: 10.1016/j.jaad.2005.10.041. Epub 2006 Jan 23. J Am Acad Dermatol. 2006. PMID: 16546580
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
